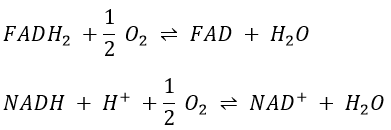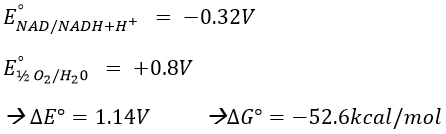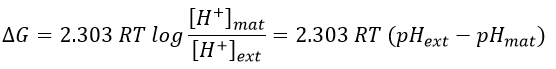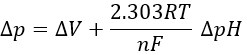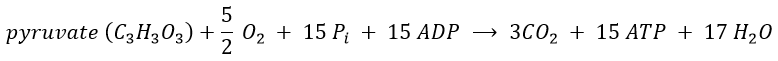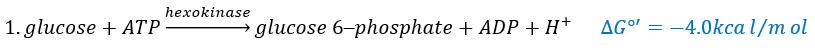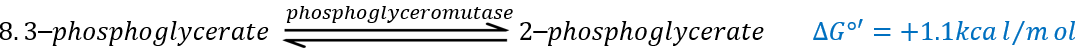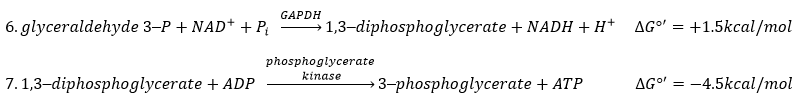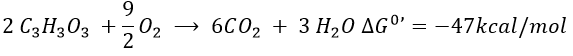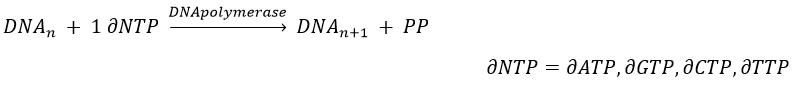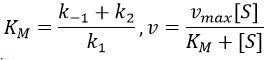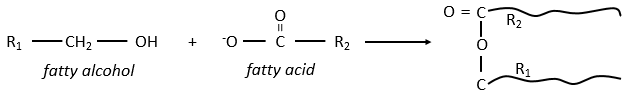Chapter 9 : Glucose catabolism – respiratory chain
In 1935, Engelhardt analysed the rate of ATP in red blood cells (globules rouges) as a function of the rate of oxygen. The experiments showed that the ATP increases with the quantity of O2. Contrarily to the yeast, the blood cells had to be in one piece to observe the phenomenon. Even more, some enzymes contained in an extract of blood cells destroy the ATP. Later, Kalkar showed that if we add one enzyme to the extract the ratio of ATP increases in presence of oxygen.
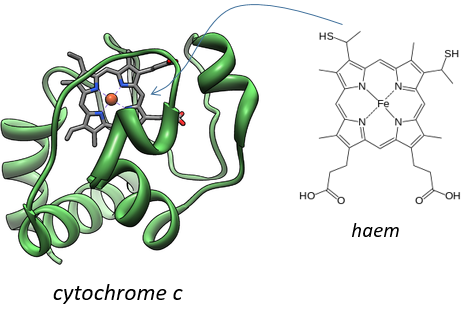
Keilin (1925) was studying the mosquitoes responsible of the malaria. With a spectromicroscope he looked at the legs of one mosquito and saw that they change of colour. At rest the muscles are oxidised but when they are excited the muscles consume the oxygen and are thus less oxidised. The colour is given by the cytochrome, a molecule that changes of colour in function of the oxidation.
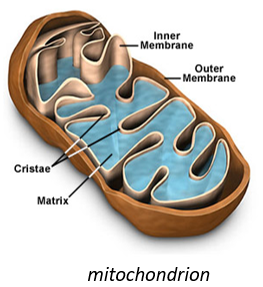
This molecule belongs to the mitochondrion. The mitochondrion is a kind of bacteria hosted by eukaryotes.
They have their own membrane, composed of an inner membrane and of a porous outer membrane. The matrix is inside the inner membrane and the intermembrane space is also called cristae where the membrane is protruding. Similarly to the haemoglobin, the cytochrome is a protein that possesses a haem the iron of which can change of oxidation state. This property is used in the respiratory chain. Cytochrome belongs to the inner membrane of the mitochondrion along which it can move and it separate the glycolysis from the citric acid cycle. The cycle of the citric acid takes place in the matrix of the mitochondrion while glycolysis takes place outside of the mitochondrion.
Respiratory chain
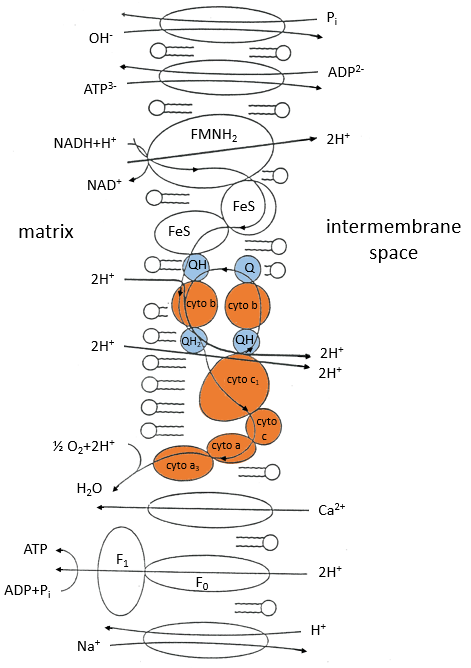
The respiratory chain can be summarised by the following figure and table:
In the inner membrane we find 4 complexes – NADH-ubiquinone reductase, succinate-ubiquinone reductase, ubiquinol-cytochrome c reductase and cytochrome oxidase – that make redox reactions and transfer electrons from one side of the membrane to the other side (sometimes protons instead of electrons). One reaction at one side of the membrane (matrix or intermembrane space) is always coupled with one reaction at the other side of the membrane. One reaction of reduction is coupled with a reduction of oxidation and vice-versa. The whole phenomenon involves transfers of electrons inside the membrane and transfers of protons from one side of the membrane to the other side. Note that some complexes belong to the membrane but only face one of its sides.
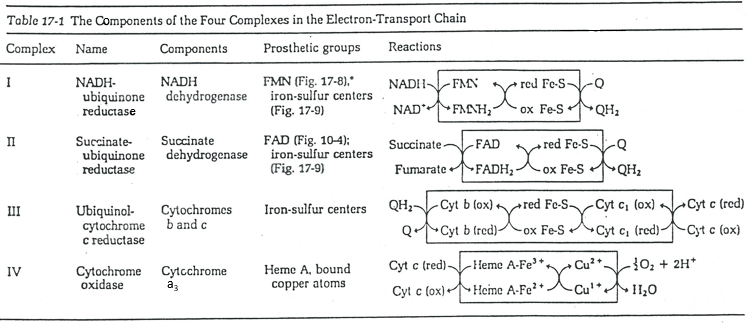
Q is the ubiquinone, a cofactor that belongs to the membrane. It interacts with the cytochrome c in a loop of oxidoreduction. One can see that the oxygen is involved by the complex IV. To resume the loop, we can write two reactions of oxidoreduction, one with the NAD and one with the FAD.
However O2 cannot directly interact with FADH2 and NADH+H+ and this loop is thus necessary. For redox, the free energy of Gibbs is
To determine the difference of potential ∆E°, we look at the couples involved in the reaction:
For FAD we have 43.3kcal/mol. There is a transfer of protons involved in the process but there must be an equilibrium between the concentrations of charge inside the cell and outside the cell. The protons can move out of the matrix of the mitochondrion through some protein complexes that generate ATP from the flow of protons (bottom of the figure). The F1-ATPase protein is composed of two parts, F0 in the membrane and F1 in the matrix of the mitochondrion. F0 pumps the protons from the intermembrane space towards the matrix. F1 receives the energy from the transport and transforms ADP into ATP. Those species are charged negatively and require a transport through the membrane (top of the figure). One ADP can go in the matrix only if one ATP is moving out of the matrix. The inorganic phosphates Pi are also transported through the membrane.
The energy to form the ATP comes from the gradient of pH. It generates a proton motive force.
Where the indexes mat and ext respectively matrix and its exterior (the intermembrane space). The gradient of pH generates a difference of electric potential ΔV.
Combining the two effects, we have
Dividing this expression by nF, we obtain the expression of a variation of free energy ∆p associated to the transfer of one mole of protons (in volt).
The coefficient 2.303RT/nF of the gradient of pH has a value of 59mV at 25°C.
We can measure the pH and the difference of potential: ∆V=0.14V and ∆pH=-1.4.
It corresponds to a variation of free energy equal to
To form one mole of ATP, we need 7.3kcal. In the respiratory chain, there are 4 complexes that oxidise the NADH and the FAD. They respectively form the equivalent of 11 and 8 protons by loop.
We can write a global equation of the “combustion” of a pyruvate during the citric acid cycle and the respiratory chain:To summarise, the oxidation of one NADH generates ~3 ATP and the oxidation of FAD generates ~2ATP.
The 15 ATP come from
- 4 NADH (one before the citric acid cycle and 3 during the cycle) à12ATP,
- from 1 FADH2 à 2ATP
- and from 1 GTP»1 ATP.
To form one pyruvate, glycolysis forms 2 ATP and uses 2 NAD that are reduced into NADH+H+.
So considering 3 ATP by NADH, it corresponds to the production of 8 ATP. Added to the 15 ATP of each pyruvate (x2), we obtain 38ATP by glucose. In comparison to the combustion of the glucose (∆G°’=-686kcal/mol), it represents about 40% of the potential energy (38×7.3kcal/mol=277kcal/mol).
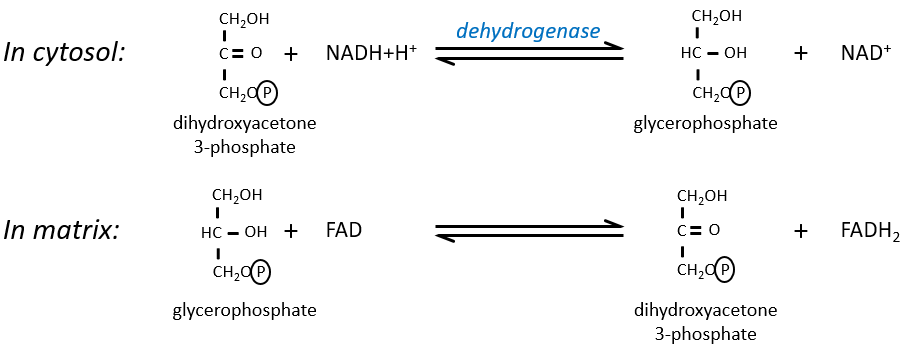
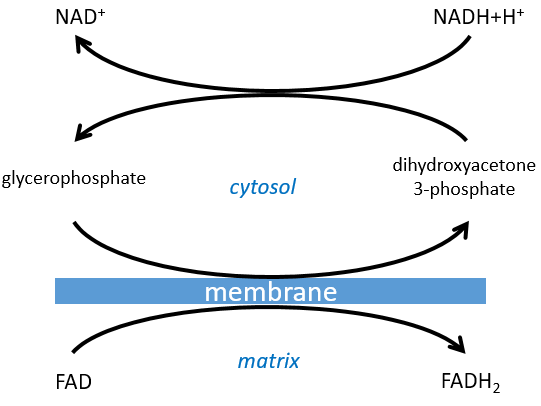
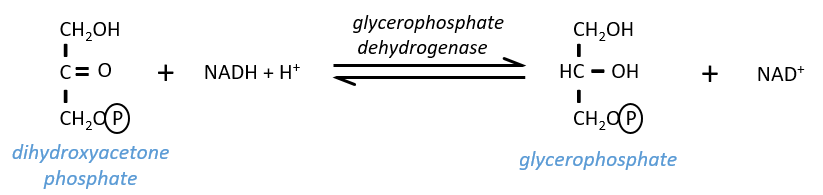
The NAD involved in the glycolysis and that produces a NADH is not at the same place (cytosol) that the NADH required in the cellular respiration (matrix of the mitochondrion). The NADH cannot enter freely into the mitochondrion because it is charged. There is a system of shuttle that may differ for different cells and that will modify the NADH to allow its passage through the membrane, then modify it again so it can be used there. In the brain and in muscles, it is a shuttle of glycophosphate that we will next explain. The shuttle is basically composed of two reactions: one in the cytosol and one in the matrix of the mitochondrion:
The shuttle can be represented like this:
If you remember, dihydroxyacetone-3P was part of the glycolysis (red rectangle in the following figure): it is one of the two trioses phosphate produced from fructose -1,6-diphosphate. The fact that this triose is involved in the shuttle does not really decrease the yield of the glycolysis significantly: the cell needs a given quantity of dihydroxyacetone for the shuttle but the molecules are regenerated in the matrix of mitochondria and can then return to the cytosol. So they just have to be produced once. The rest of the production is turned into glyceraldehyde 3P for the glycolysis.
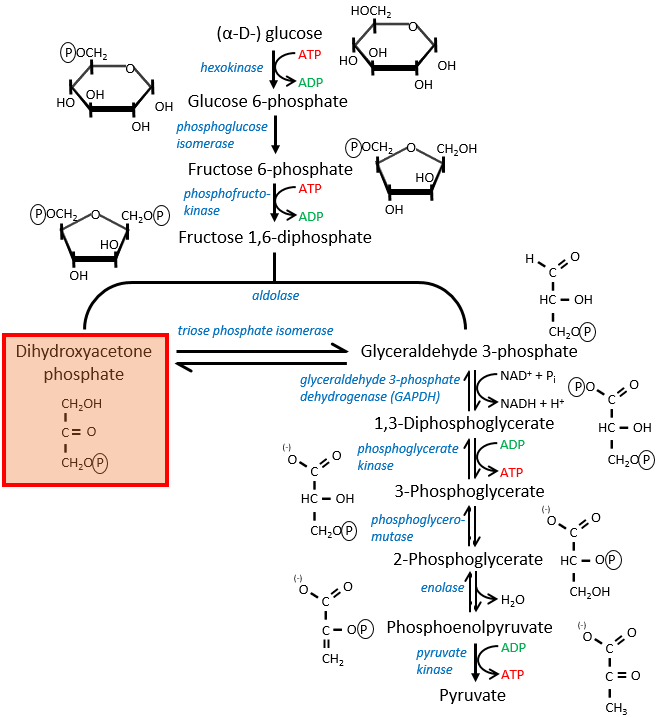
The FADH2 is oxidised during the respiration and the dihydroxyacetone can move back to the cytosol. The shuttle has a small cost: it oxidises one NADH into NAD in the cytosol while the reaction in the matrix involves FAD/FADH2. In term of energy, it represents one ATP used because all the processes don’t take place at the same place.
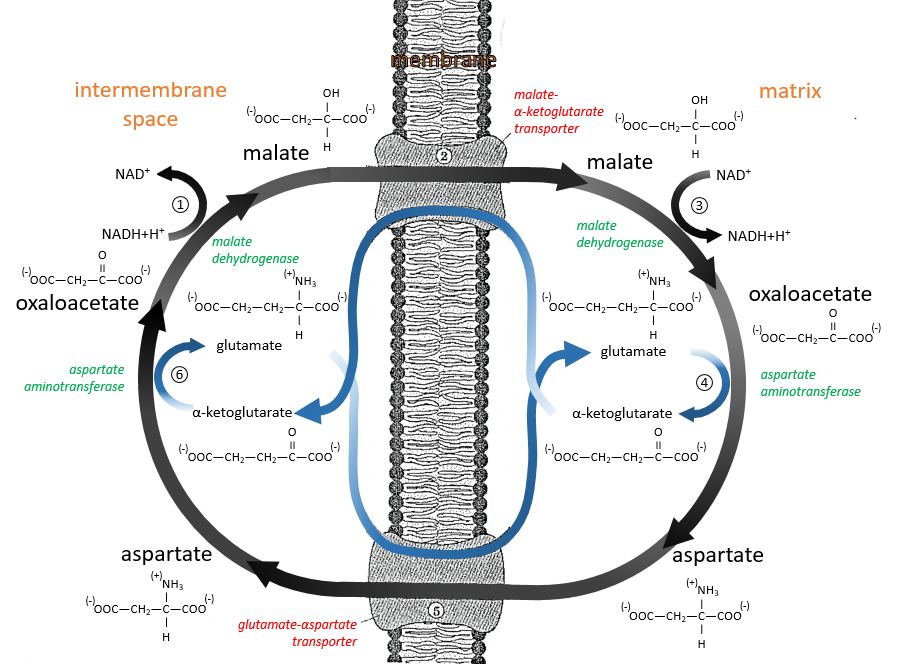
The malate-aspartate shuttle
The same cofactors (NADH+H+/NAD+) are used so the yield does not change with this shuttle (38 ATP). The mechanism is based on the reducing power of the oxaloacetate.
Reduced into malate (1), it is transported in the matrix of the mitochondrion (2) through the malate-α-ketoglutarate transporter if one α-ketoglutarate is available to make the displacement in the opposite direction. The reverse reaction, the oxidation of the malate into the oxaloacetate is made in the matrix (3). Yet, the oxaloacetate cannot goes back out of the membrane. To do so, it is transformed into an amino acid, the aspartate, by a reaction of transamination (4):
It is done here by the aspartate transaminase with the use of the glutamate as amino acid. As a result, the oxaloacetate is transformed into aspartate that can move through the membrane through a transporter requiring the passage in the opposite direction of a glutamate (5). In the cytosol, it is turned back into the oxaloacetate with the exact same reaction (6) than at the other side of the membrane.
Chapter 8 : Glucose catabolism – aerobic oxidation
This process is coupled to the cellular respiration, involves O2 and is way more effective than the anaerobic oxidation. Instead of 2 ATP, the aerobic oxidation generates 38 ATP by glucose. It can also oxidise fatty acids and the carbonate parts of amino acids.
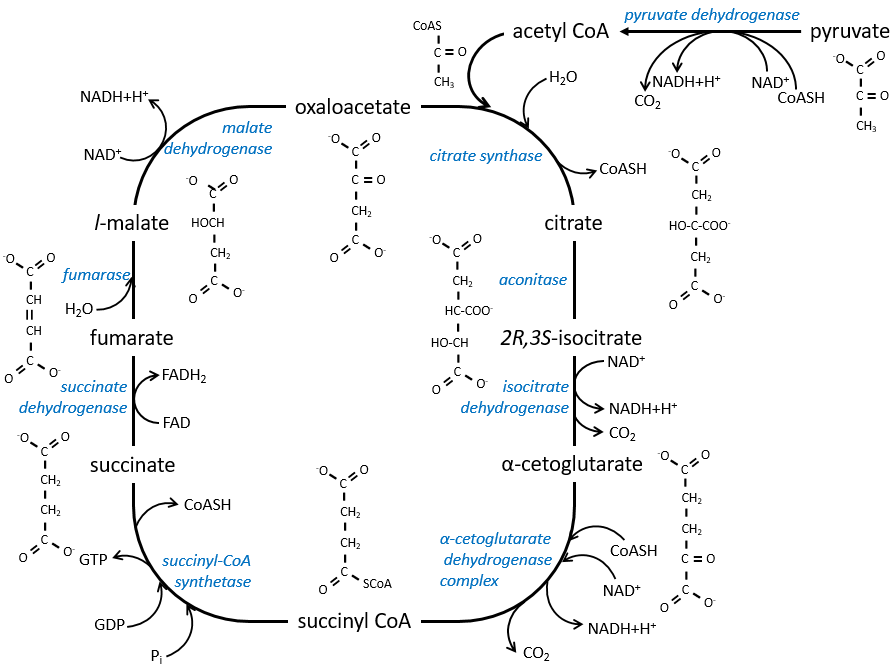
The cycle of Krebs: cycle of the citric acid
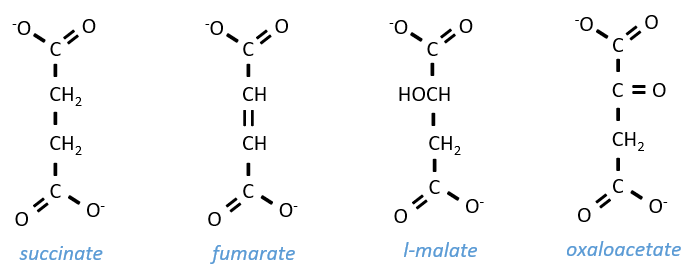
Szent Fuorgue studied the cellular respiration from an extract of muscle. He analysed the absorption of O2 in presence of various molecules. He observed that some of them considerably increase the absorption of oxygen by the tissues. It is the case for the succinate and the fumarate.
These molecule are stable to variations of temperature. The combustion of one succinate normally consumes 3 O2 but during the experiments they observed that the absorption of oxygen was above this value. They also showed that the absorption can be decreased if we add malate to the system. The malate is an inhibitor that affects the oxaloacetate, one catalyst of the cycle of Kerbs that is regenerated at each cycle except in presence of malate.
The cycle can be resumed by the following figure.
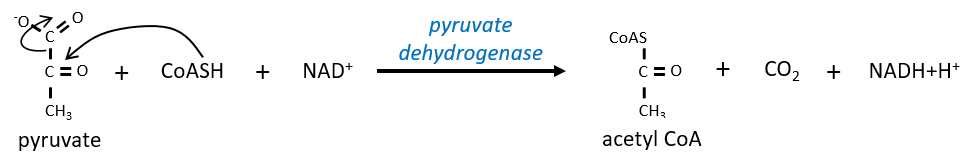
Citrate is the product of the first reaction of the cycle. Before explanations on the cycle, we will explain the formation of the acetyl CoA. It is made from pyruvate. The first step is the substitution of one CO2 by the CoASH through a thioester liaison.
The reaction is irreversible (release of CO2) and is catalysed by a big enzyme: the pyruvate dehydrogenase. Inside the enzyme we find the B1 vitamin. Simultaneously, there is an oxidation made by the NAD+. The product is the acetyl coenzyme A, or acetyl-coA.
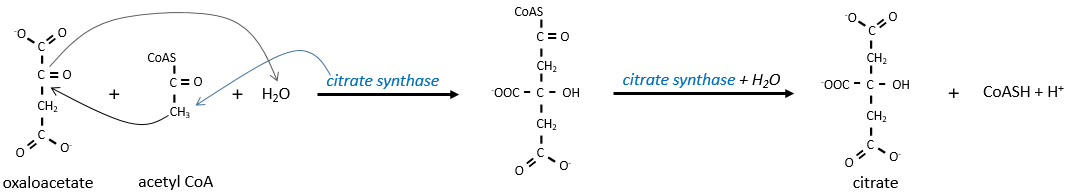
The first step forms the citrate by the condensation between the acetyl-coA and the oxaloacetate. The reaction is helped by the citrate synthase that take a proton from the methyl group of the acetylCoA. It releases the coenzyme that can be used again.
The citrate is symmetrical but it is considered as prochiral by the enzyme of the next reaction. To pursue to cycle, the aconitase changes the conformation of the citrate to obtain the isocitrate.
The equilibrium is heavily in favour of the left but the right species is consumed by the next reactions so the reaction is displaced towards the right. Step 3 is subdivided into one reaction of oxidation and one decarboxylation.
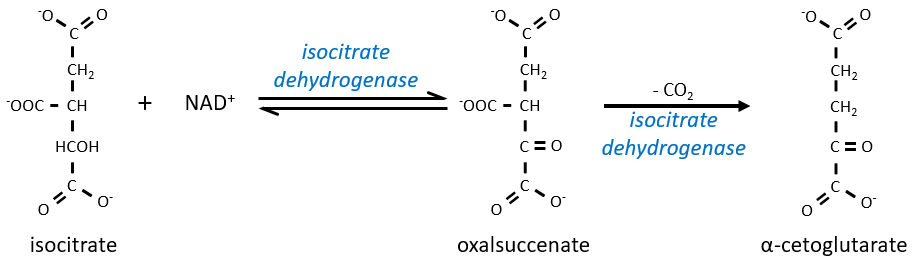
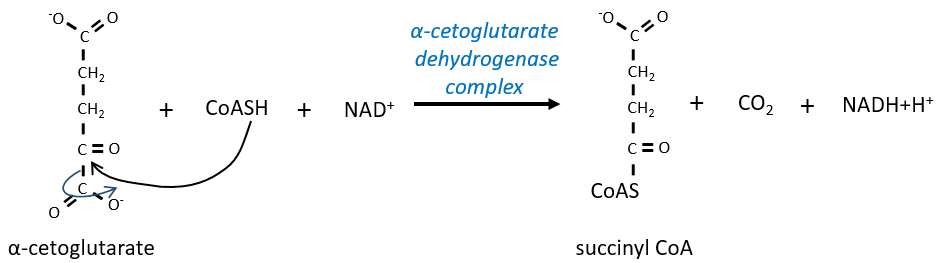
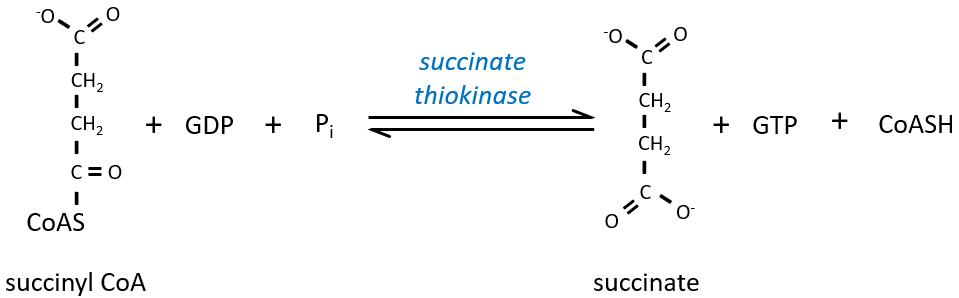
The step 4 is similar to the formation of the acetyl-CoA and leads to the formation of the succinyl-coA.
The cleavage of the thioester liaison gives enough energy to form, not an ATP but a GTP.
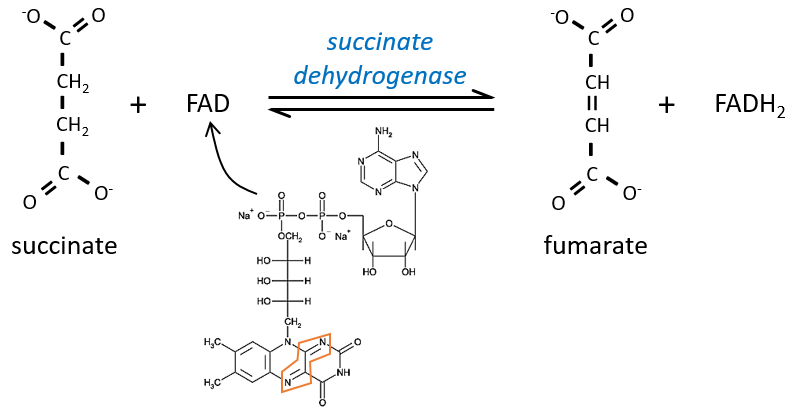
The succinate is next oxidised to for the fumarate. The cofactor is the FAD: Flavin adenine dinucleotide, a derivate of the riboflavin (vitamin B2).
The reduction of the FAD is done at the orange area.
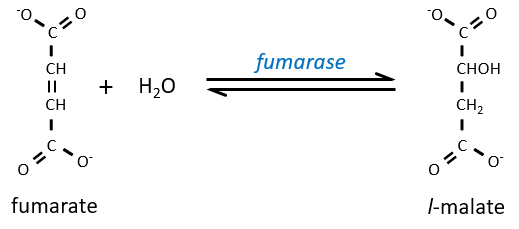
Step 7 needs one molecule of water to form the malate. Malate is a chiral molecule, the chirality given by the structure of the active site of the enzyme. As a result, only the l-malate is obtained.
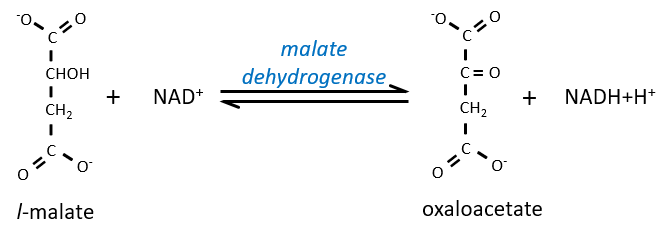
Finally, the oxaloacetate is regenerated from the malate.
Now, you may say that there is none of the 38 ATP promised at the beginning of the section. Only one GTP was formed during the cycle but several CO2 were rejected and 3 NAD+ and one FAD were reduced. These molecules represent the energy of the ATP. Moreover, it is the process that occurs in aerobic cells but no O2 was ever involved in the cycle. We will see the source of the ATP in the next section: the respiratory chain.
Chapter 7 : Glucose catabolism
Its goal is to supply energy to the cell wherever it is needed. The glycolysis forms pyruvate from the glucose that can next be deteriorated anaerobically to form lactate or ethanol through fermentation. In 1870, Louis Pasteur discovered the functioning of yeasts. He isolated one yeast and added it to a wine that was not fermenting. The wine fermented. At this time, Pasteur believed in the vitalism so he did not search further. Later, the Buckner brothers were preparing extracts of yeast. As it was not easy to conserve, they put the extracts into sugar and it fermented. The yeast has thus not to be complete (or alive) to be effective. The extracts were composed of two molecules: one large enzyme that becomes ineffective when the temperature increases and one small molecule that was ineffective alone. In fact, the small molecule is a coenzyme of the big enzyme and the fermentation is only possible when both molecules are together. The complete process of the fermentation of the glucose was discovered in 1925 by Embdem and Meyerhof.
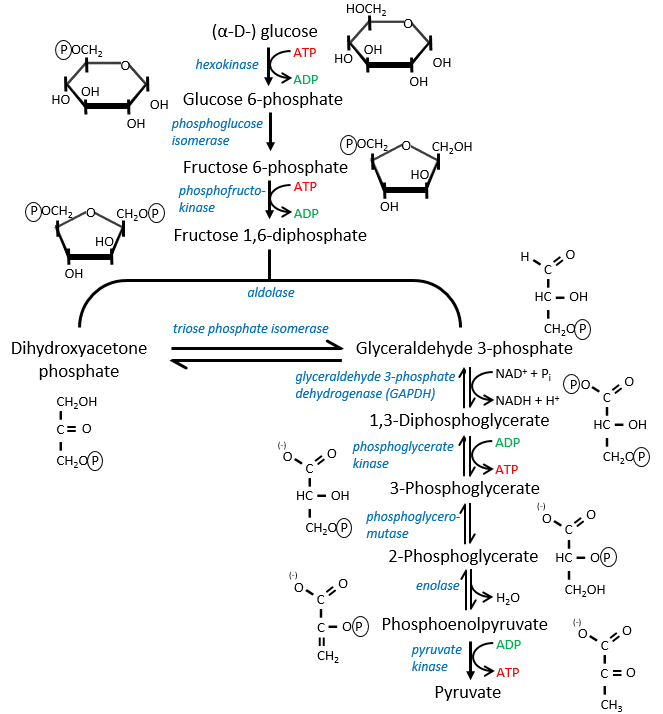
The first step is the phosphorylation of the glucose. The phosphate is added on 6’. For this reaction, as for the others of the glycolysis, an enzyme (the hexokinase/glucokinase) to form the ester and to cleave the anhydride of the ATP.
The second reaction involves the transformation of a pyranose into a furanose.
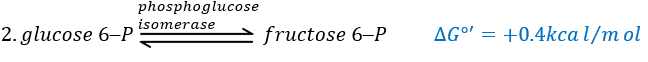
The third step is dependant of the needs of the cell in energy.
![]()
When the cell absolutely needs energy, its consume ADP and forms AMP. If the concentration of AMP in the cell increases then a signal is sent to the phosphofructokinase to increase the rate of the reaction. On the contrary, if the cell is full of ATP, then there is no need to make more of them and the reaction is slowed.
![]()
In the next step, the fructose is cleaved into two chains of 3 carbons, an aldose and a ketone that can be transformed into each other by the triose phosphate isomerase.
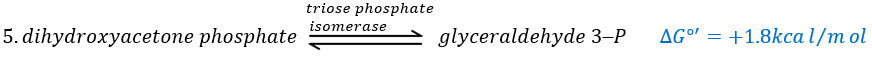
Because the reaction 5 is reversible, if the C1 of the glyceraldehyde was marked, we would find the fructose marked on 3’ or in 4’.
The next steps involve chains of 3 carbons. From one glucose, they are thus occurring twice.
![]()
The enzyme of the step 6 is the GAPDH (glyceraldehyde 3-phosphate dehydrogenase) and the NAD (nicotinamide adenine dinucleotide) is its cofactor and has to be in the active site of the enzyme for it to work. NAD+ is its oxidised form and NADH+H+ its reduced form.

The presence of the reduced form can be confirmed by a new peak of absorbance at 340nm. The reduction generates some energy to allow the formation of a phosphoanhydride, what is a liaison high in energy. It will thus generate a lot of energy when it is cleaved.
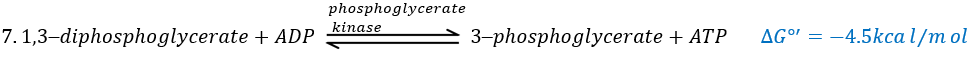
The seventh step uses the energy of this liaison to regenerate one ATP.
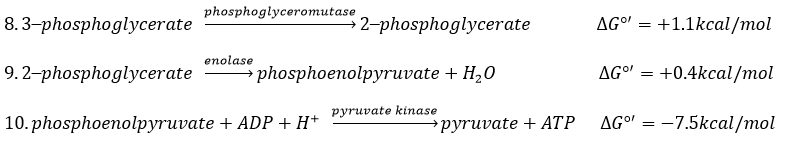
The eighth reaction displaces one phosphate. It is done by an enzyme called mutase.
![]()
The ninth step is an intramolecular reaction of oxidation. C2 is oxidised while C3 is reduced. We have now a phosphoenol the cleavage of which restores one ATP in the following reaction of tautomerization.
![]()
In total, two ATP were used during the glycolysis to form the trioses. These steps are thus using the reserve of energy of the cell. However two reactions form one ATP by triose. As there are two trioses by glucose and the isomerase can turn the ketone into the glyceraldehyde, so we form 4 ATP from the trioses. During the complete glycolysis, 2 ATP are thus formed that can be used by the cell. For anaerobic cells, it is the only way to produce ATP.
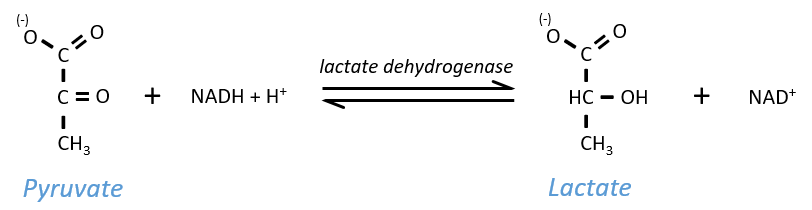
The NAD+ has to be regenerated by fermentation.
Lactic acid fermentation
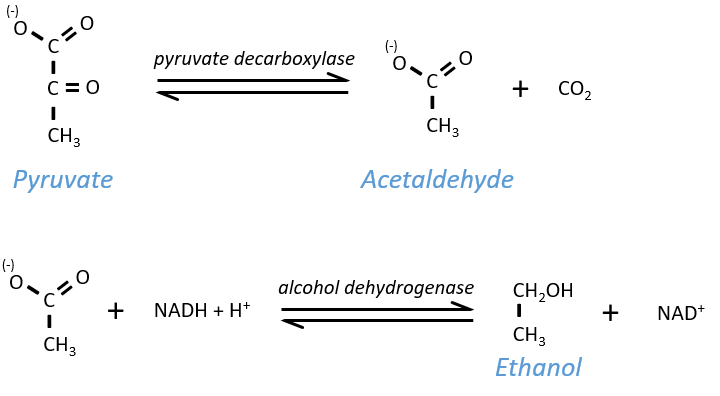
The pyruvate, the final product of the glycolysis, is used to oxidise the NADH+H+ and form lactate that is next released in the middle.
Ethanol fermentation
Also called alcoholic fermentation, this process also uses the pyruvate but is made in two steps. One first that involves the rejection of CO2 and the second that oxidises the NADH+H+ and frees some ethanol.
Comments on the glycolysis
- In the case of a lack of NAD+, or a too large quantity of glucose, more NAD+ can be formed from the “unused” triose, the dihydroxyacetone phosphate
The reaction is in equilibrium and is only made in case of need. If the triose was essentially used to do this, the whole process of the glycolysis would be pointless: it would consume 2 ATP to form 2 ATP.
- Coupling of reactions
Some steps of the glycolysis have a positive ∆G0’. It is the case of the steps that form rich liaisons. In this way, the step 6 is coupled to the step 7 that cleaves the rich liaison to form an ATP.
If we consider the two reactions at once, i.e. a reaction of oxidation of the aldehyde into a carboxylic acid, we find a ∆G0’=-10kcal (7.3kcal are equivalent to one ATP). The two-steps reaction is favoured because we can store the energy as ATP.
The same is true for the steps 8, 9, 10.
The two first steps cost energy but the third is very favourable. In total, we have a ∆G0’=-6kcal/mol and the formation of an ATP.
- Uncoupling agent
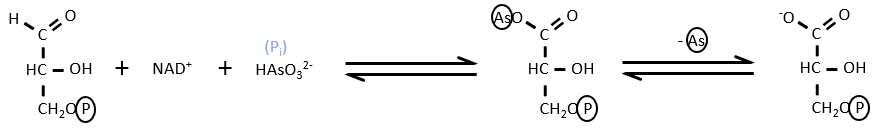
There are some molecules that can block the formation of the ATP. It is for instance the case of the arsenate ion which is a poison for us. In the step 6 and 7, the ion takes the place of an inorganic phosphate.
We still obtain the product of the step 7 but without the formation of ATP.
- energetic balance
The combustion of one glucose is an exothermic reaction:
The combustion of the products of the glycolysis (two pyruvates) gives
It is about 7% of the energy of one glucose. To that we can add the energy of the phosphoester bond of the 2ATP: 2×7.3kcal/mol.
It is still a very small quantity of energy that was recovered from the glucose. This method was used when there was almost no oxygen available and is used in anaerobic environments.
- Other monosaccharides can make the glycolysis after being transformed by enzymes into glucose or one intermediate of the glycolysis.
Chapter 6 : nucleic acids
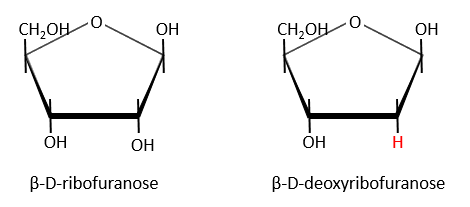
Nucleic acids are composed of monosaccharides connected by phosphoester liaisons and wearing a base. One monosaccharide and its base are called one nucleoside. One monosaccharide, its base and one phosphate are called a nucleotide. RNA and DNA are composed of one similar monosaccharide, the ribofuranose, with the difference that the 2’ has an H on the DNA in place of an OH for the RNA. It is the reason of their names: the ribonucleic acid (RNA) and the deoxyribonucleic acid (DNA).
Monosaccharides are bound together in 3’-5’ and the base is bound by the OH on 1’. There is a total of 5 bases.
- purine bases: they are composed of two aromatic cycles with nitrogen atoms in the cycles. They absorb the UV at 260nm.
- adenine (A)
- guanine (G)
- pyrimidine bases: they are composed of one aromatic cycle and also absorb UV.
- uracil (U, only found in RNA)
- thymine (T, only found in DNA)
- cytosine (C)
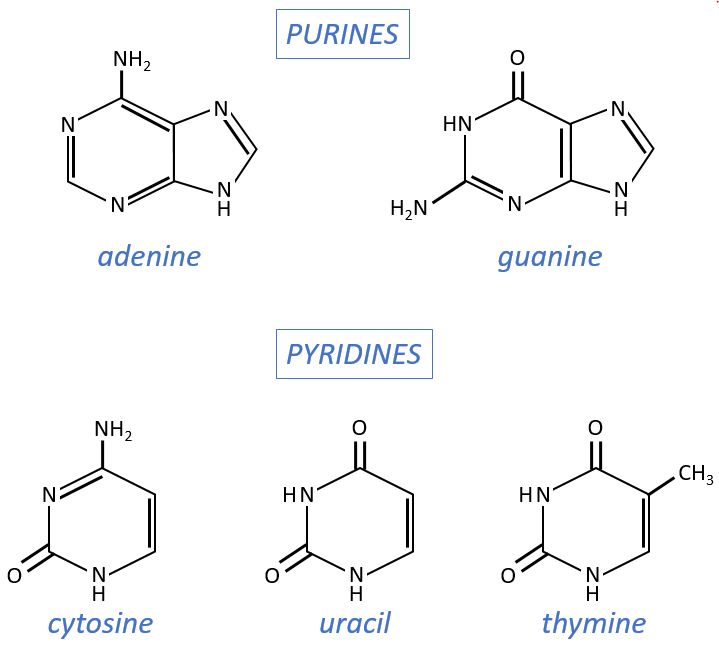
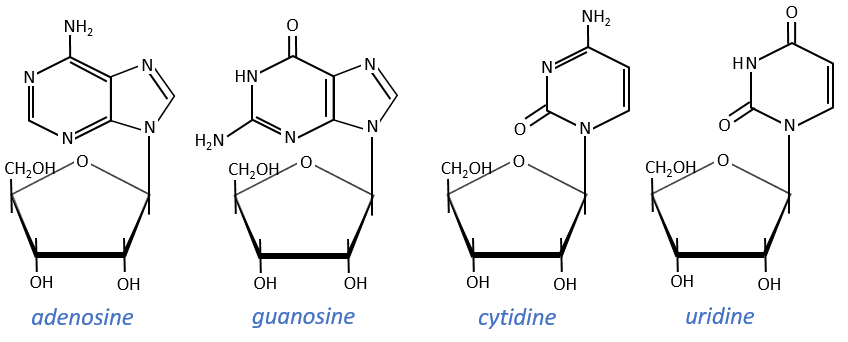
Nucleosides are the combination of one base with one monosaccharide. The names of nucleoside are directly related to their base.
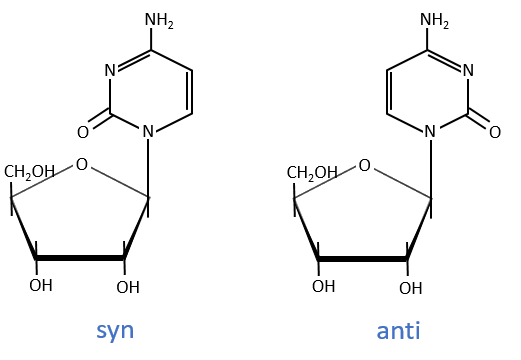
The same is true for the deoxynucleosides. The prefix deoxy is just added to the name of the corresponding nucleoside. There is a stereochemistry in the orientation of the base and the monosaccharide. The liaison can either be in anti or in syn.
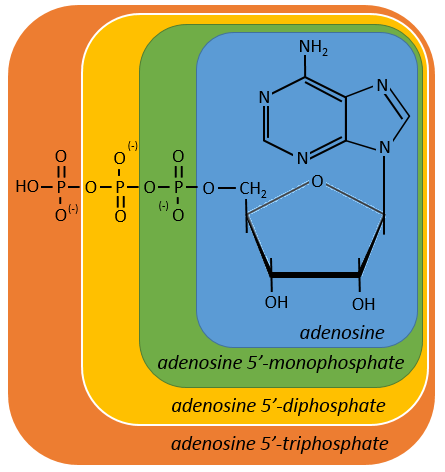
Nucleotides are the combination of one nucleoside with one or more phosphates (in chains). The phosphates can be placed on 5’ or on 3’ by an ester liaison and phosphates are bound as acid anhydride of acid. The liaison between two nucleosides is a phosphodiester liaison.
One example: two important nucleotides are the adenosine diphosphate (ADP) and the adenosine triphosphate (ATP).
If the base was a deoxy~, we write a ∂ (or d) before the name (ex: ∂ATP)
Nucleotides absorb in the UV the same way nucleosides do.
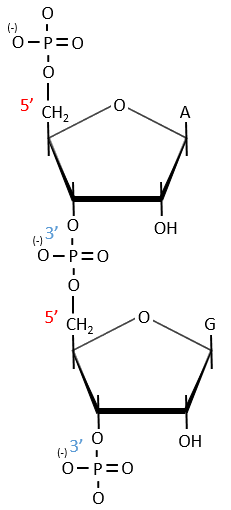
A chain of nucleic acids has a polarity. It goes from the 5’ towards the 3’. A chain is written in this order in abbreviated:
The chains are also build following this order.
Cleavage of the chain
- By total acid hydrolysis, using a 12M HCl solution. It cleaves all the liaisons between the monosaccharides, the bases and the phosphates.
It is the harsh method, the following methods are gentler.
- By depurination: we work at pH=1.6 and at 37°C. The cleavage only works for the DNA and only break β-N-glycosidic bonds. β-N-glycosidic bonds are hydrolytically cleaved releasing a nucleic base, adenine or guanine, respectively.
- hydrolysis for RNA: we use NAOH 0.1M at 25°C (during 15hours). It only cleaves the RNA chains because the reaction involves an intermediate species that needs the OH in 3’
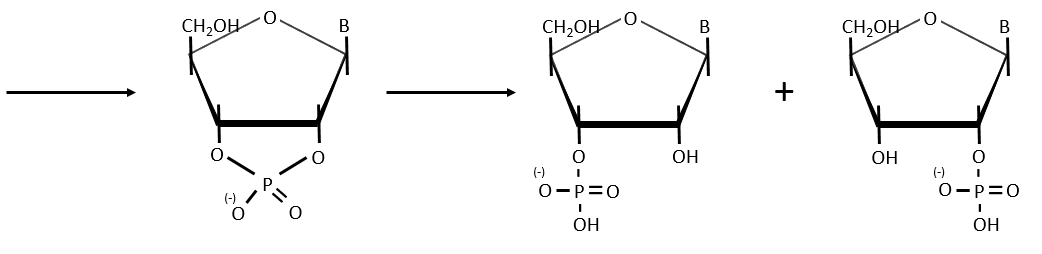
- enzymatic cleavages: some enzymes are specialised in the cleavage of nucleic chains. There are of two kinds: endonucleases and exonucleases. The firsts cleave liaisons inside the chain and the seconds cleave the chains starting at one extremity. It can go from 3’ to 5’ or from 5’ to 3’, depending on the enzyme. The extremity has to be free of phosphate to be cleaved. For instance, the phosphodiesterase that we have in the spleen only works if the 5’ extremity is ended by OH. It will free nucleosides 3’-phosphate. The phosphodiesterase that we find in the venom of snakes cleaves the 3’ extremity and forms nucleosides 5’-phosphate.
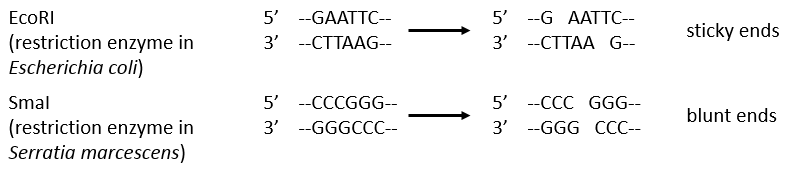
Endonucleases are usually not specific and give nucleosides 3’ phosphates. Yet there are some specific endonucleases that recognise a particular pattern in the chain. For instance, endonucleases of restriction are enzymes that we find in bacteria’s to protect themselves from bacteriophages. Bacteriophages are viruses that attack bacteria’s and try to inject their own DNA inside the DNA chain of the bacteria. The enzyme recognise a palindrome in the sequence of the DNA. It is called a site of restriction. This site is cleaved in a special way.
The resulting extremities may be sticky or blunt, depending if the cleavage is done at the same base or not on the two strands. The term sticky was chosen because they are easily joined back together by a ligase if there are several unpaired nucleotides.
The DNA of the bacteria is protected from this cleavage because its DNA was methylated by a methyltransferase after/during its formation.
Structure of the DNA
DNA has a helical structure that was determined by several experiments.
- Chayaff: he was studying the composition of the DNA in term of bases. He performed a total acid hydrolysis and analysed then the proportions of A, C, T and G. The result of its experiment was that the proportion of A is equal to the one of T and that the proportion of G was equal to the proportion of C. Moreover, the sum A+G=C+T.
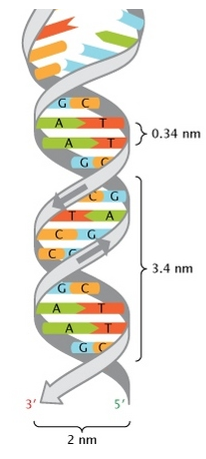
- Wilkin and Franklin: They performed the XR diffraction of the DNA. They observed particular pattern with a two characteristic lengths of 0.34nm and of 3.4nm.
- Watson and Crick: From the experiment of Wilkin and Franklin, they gave a model of structure for the DNA: a double helix.
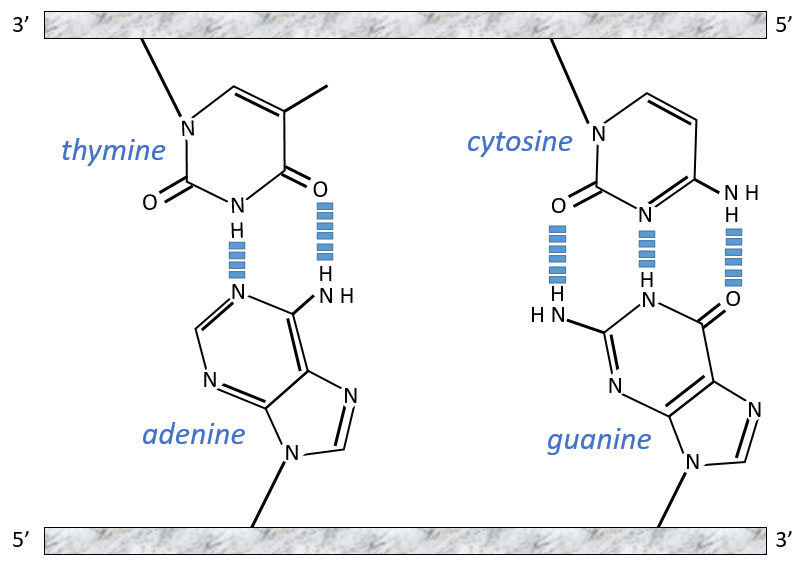
The chains of monosaccharides are at the outside of the double helix and the bases are facing each other at the inside of the helix. Each base can only be pairing one other base: A with T and C with G. It explains the equivalences in proportion.
The helix is stable thanks to the H bonds between the bases. There are 2 H bonds between A and T and 3 between C and T.
As they are surrounded by the hydrophobic chain of monosaccharides, the bases are protected from water that could interact with the H bonds linking the bases. There are also electrostatic interaction between the bases.
A heating (80-90°C) can separate the two “strands” of DNA from each other. When separated, they absorb more light than when they are coupled and we can observe a hyperchromic effect at 260nm.
The sites where enzymes cleave the DNA are always full of A or/and T. It is easier to open the helix here because there are less H bonds between the bases.
The length of the DNA varies between species and is not in helix for the prokaryotes for which the DNA is circular. The longest genome belongs to the “triton”
Replication of the DNA
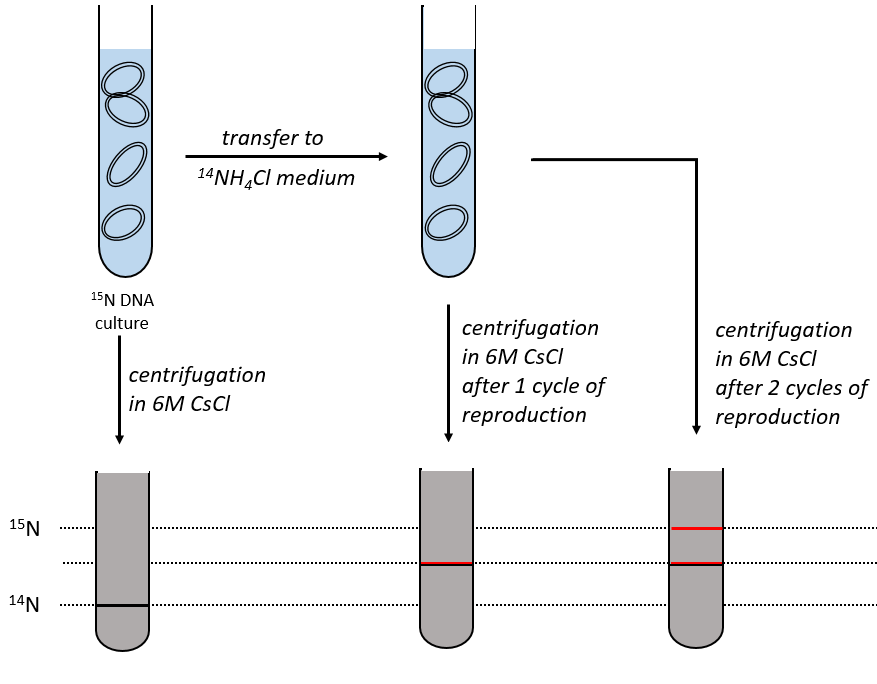
During the mitosis, the cell splits in two cells, the DNA is reproduced and each new cell possesses the same genome than the cell they are made of, the mother cell. The DNA can spontaneously reform itself if it was denatured as it was shown by the experiment of Miseber and Slahl: they placed some bacteria’s in a solution the composition of which is controlled: the single source of nitrogen in the solution is NH4Cl and the nitrogen were marked isotopes. The bacteria’s form the bases of the DNA with this nitrogen. After a while, the DNA is centrifuged with a dense salt of CsCl. During the centrifugation, the salt goes to the bottom of the tube and the DNA stops at its density. As there are two isotopes of nitrogen (14N and 15N), there will be two layers of DNA with different densities.
We can thus isolate the light DNA from the heavy DNA. If we place them respectively in a solution of culture with 14N and with 15N, they can replicate. From the solutions, we extract some DNA, put them together and denature them (T=100°C). The strands of DNA are thus separated. When the solution is cooled down, the strands pair together but not necessarily with the strand they were paired with previously. After centrifugation, there are still the layers of DNA that were obtained previously but there is now a third layer between the two other, corresponding to the binding of one 14N strand with one 15N strand.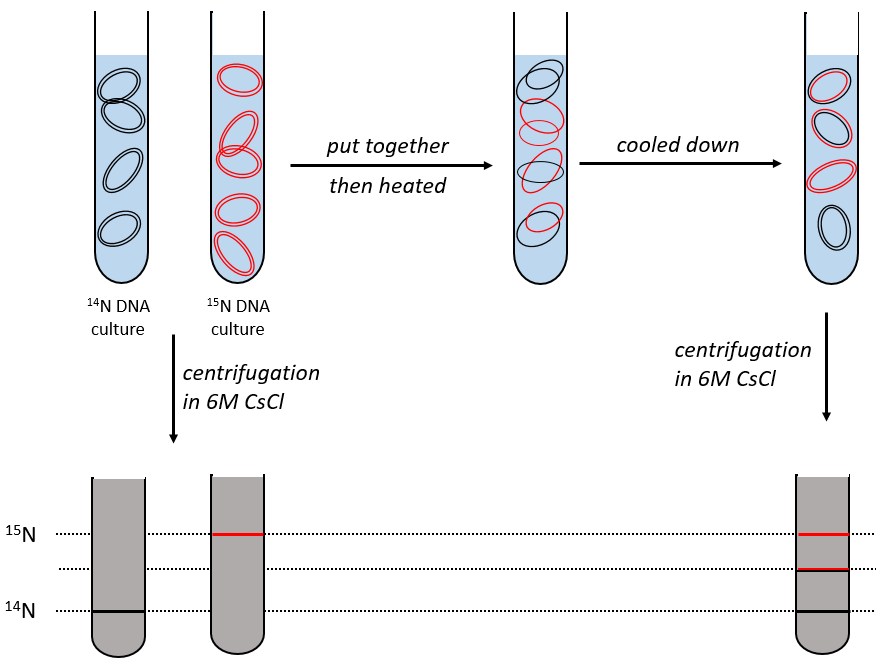
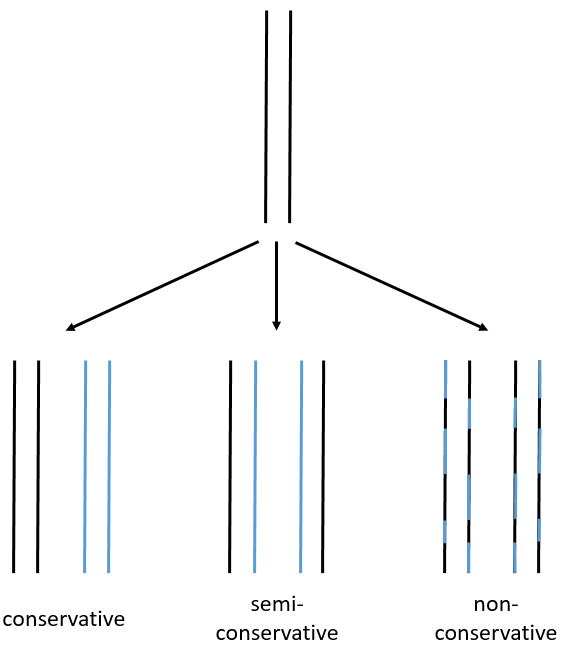
We can imagine three ways of replication for the DNA: we consider a mother strand of DNA and one daughter strand that is the replication of the mother strand.
- conservative replication: the two strands of the mother DNA remain paired after the replication.
- semi-conservative replication: one strand of the mother DNA pairs with one strand of daughter DNA.
- non-conservative replication: the sequences of mother and daughter DNA are mixed in both strands.
The replication of the DNA is semi-conservative. It can be confirmed by placing the 15DNA in a culture with 14N. If the replication was conservative, we would only obtain two layers of different densities. If the replication was non-conservative, there would be a continuous layer of DNA in the centrifugation tube. After one replication, there is only one layer (mix of 14N and of 15N). After two replication, there are two layers in the tube (14-15N and 14N). It confirms that the replication is semi-conservative.
The replication of the DNA is done by the DNApolymerase, which takes one strand of DNA as model and replicates it from 5’ à 3’, adding one nucleotide by one. This addition is done from deoxynucleosides triphosphate and generate an inorganic phosphate (or pyrophosphate)
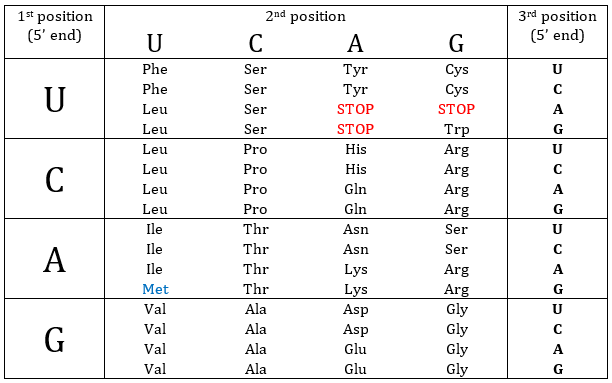
Nucleotides form triplets, called codons. The table below shows the codons and the names we call them with.
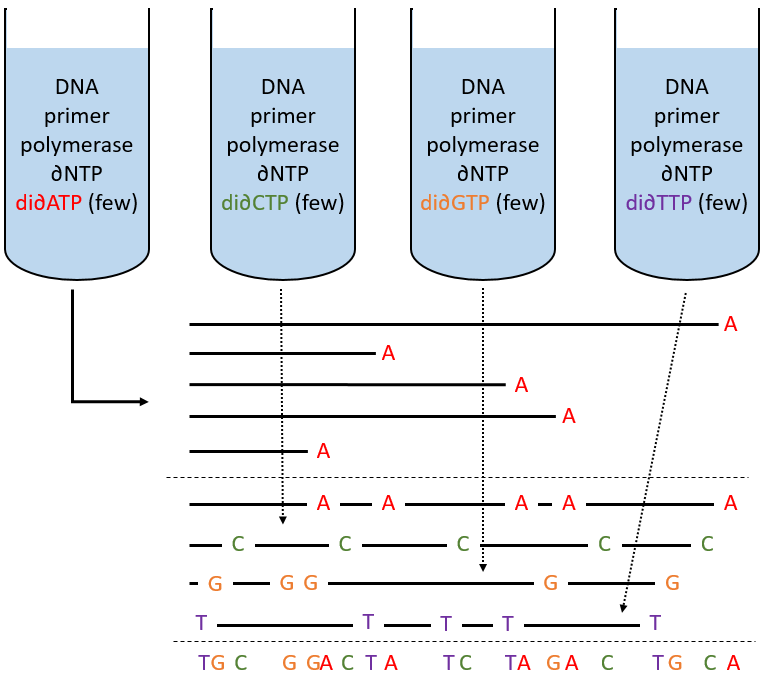
Determination of the sequence: the method of Sanger
To use this method, we need a complete strand of DNA and one fragment of it obtained via an endonuclease. The fragment is called a primer. We also need to prepare 4 different solutions. In each there are some DNA polymerases and some of each ∂NTP. One of the ∂NTP is marked. In the first solution, we add a few di∂ATP. In this dideoxynucleotide, the OH of the 3’ is missing and it stops the chain. In the second solution, we place a few di∂CTP, in the third a few di∂GTP and in the fourth a few di∂TTP. When we put the primer and the DNA in one solution, the DNApolymerase synthesises a strand at the extremity of the primer, copying the sequence of the DNA. However the synthesis will stop when one di∂NTP is added to the sequence. In each solution we know which nucleotide ends the sequence but the chains can differ in length: for instance in the first solution if one adenosine was to be added by the DNApolymerase, it can come from a ∂ATP or from a di∂ATP. In the first case the chain continues to grow and not in the second case.
The same happens in the other solutions except that the strand ends by a different nucleotide. We analyse the 4 solutions in parallel on an electrophoresis gel. On this gel, the smallest molecules move faster than the long ones and they can be recognised via to the marked nucleotide. We can thus determine the size of each fragment knowing its final extremity. As a result, the complete sequence can eventually be determined.
Chapter 5 : 3D structure of proteins
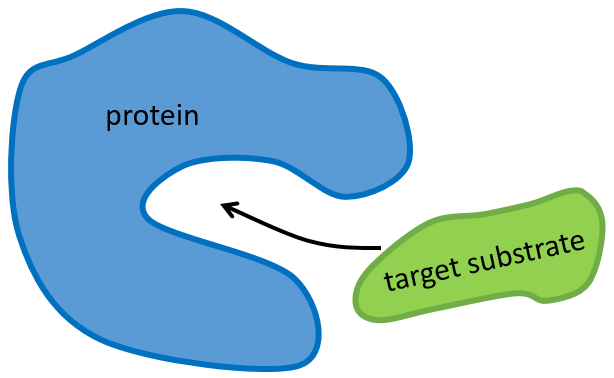
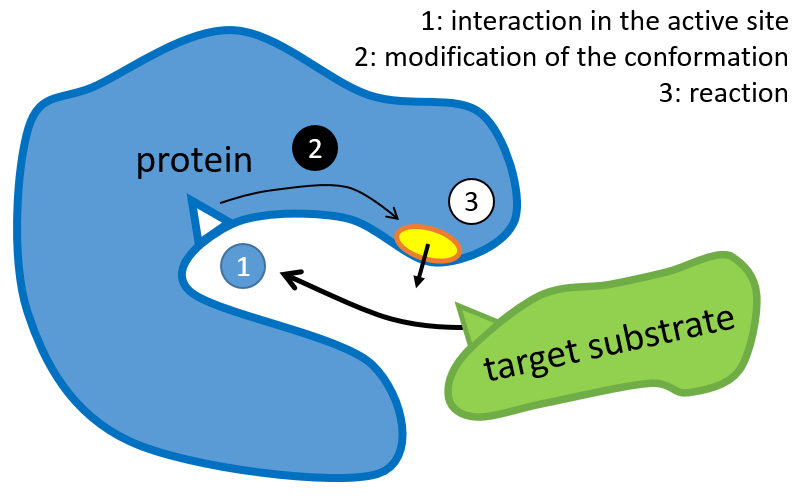
The structure of the protein is obtained during the formation of the protein. The structure of the protein is necessary for its function: some active sites are present on the proteins. Those sites are highly specific to one target substrate. Other substrates cannot reach the active site.
The rest of the molecule is mostly there to ensure the specificity of the reaction or a modification of the conformation of the protein once an active site is activated (to submit a signal for instance). The peptide chain can also help the reaction to occur by the application of a pressure on one specific liaison of the target molecule.
A protein is denaturised when its conformation is modified. Consequently, its function is lost. It can be done through heating (~60°C), a strong base/acid, … It is frequent that under those conditions the protein precipitates.
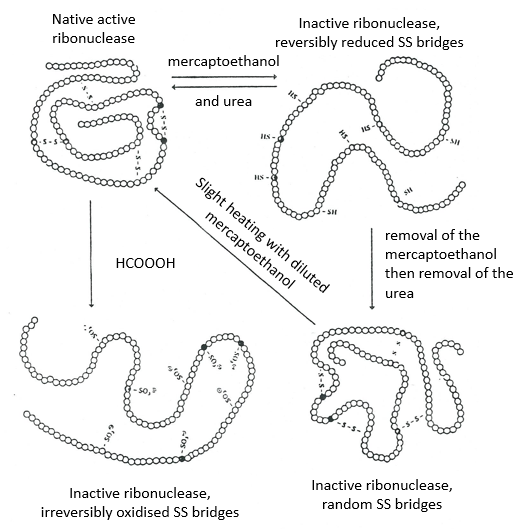
The determination of the spatial configuration of the ribonuclease was done in 1950.
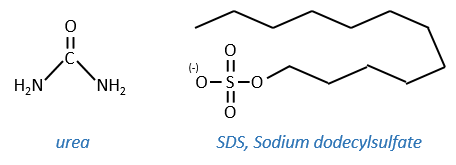
This protein has 4 disulphuric bonds (8 cysteines). Anfinsin placed the protein in a solution with mercaptoethanol and urea (NH2-CO-NH2). The mercaptoethanol cleaves the S-S bonds while the urea forms H bonds with the protein. The activity of the protein decreased by 99%. The normal conformation of the protein could be reobtained by placing the solution is a dialyse bag: placing the bag in a clear solution without urea and without mercaptoethanol, these molecules move out of the bag while the protein remains inside. Its activity comes back to 100%. It is interesting to note that the S bonds were all formed at the right place (amongst 105 possibilities). The mechanism is too fast to be try and errors.
If we removed only the mercaptoethanol (the solution outside the dialyse bag has already urea in it), the S bonds are done but at the wrong places.
All the proteins don’t get back to their functional form after a denaturation. They are build sequentially during their synthesis and the interactions between the chains are done before the complete synthesis of the protein. It is thus unusual that the sequence that was made first binds with one of the last sequence. However, after a denaturation, any part of the peptide chain can bind with any sequence of the chain, no matter the order of formation.
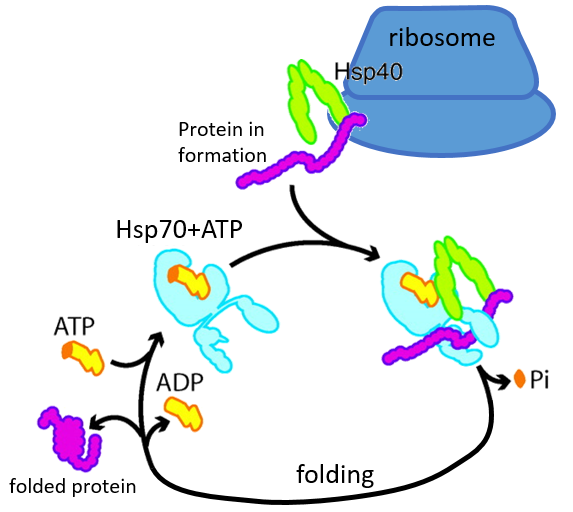
Some proteins are there to help to the correct deployment of other proteins. They are called heat stock proteins (hsp70 and hsp60): after an increase of temperature, several hsp70 recognizes the hydrophobic parts of peptide chains and bind with the protein. It helps the protein to reach the correct conformation and the hsp70 leave the protein next. The hsp60 acts on proteins that are already wrongly deployed and correct their structure.
Determination of the structure of proteins
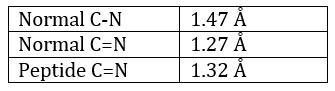
The method is based on the diffraction of X rays. From crystals of the proteins we obtain a distribution of the atoms. Payling and Corey analysed the interatomic distances between the different atoms in the amino acids. They found out that the peptide bond has a length between the length of a liaison C-N and a liaison C=N.
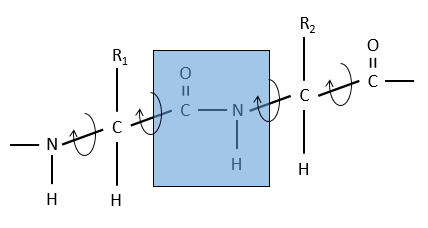
The reason is the resonance with the carbonyl. The peptide bond is thus rigid, almost always trans, but the liaisons around the peptide bond can rotate.
Secondary structure
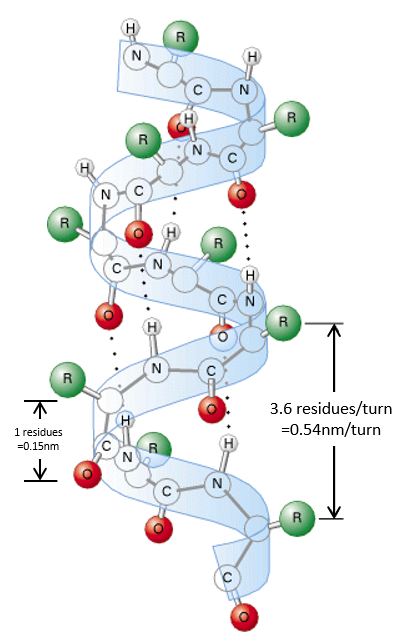
The secondary structures of a peptide chain are small “motifs” due to the interaction between nearby AA. The first structure, the α helix, was proposed in 1951. The distance between spires of the helix is 0.54nm and the length of one AA is 0.15nm. There are thus approximatively 3.6 AA by spire. All the R groups are at the outside of the helix, which is always L. There are H bonds between AA distant of 3-4 AA
All the AA cannot take place in helices. The structure is compact and AA with voluminous R group (for instance with an aromatic in R) would deform the helix.
The proline cannot be part of a helix too: its R is covalently bound to the nitrogen of the peptide bond. The liaison is thus particularly rigid and cannot be placed in one helix.
A third exclusion of the helices is charged amino acids. Imagine a helix with several –COOH groups. The structure is stable in neutral conditions but if the pH changes, there can be a lot of nearby COO-, what is unstable.
We find a lot of α helices in fibrous proteins such as the keratin α (found in the hairs). The hydrophobic AA form helices and we find them in lipid membranes.
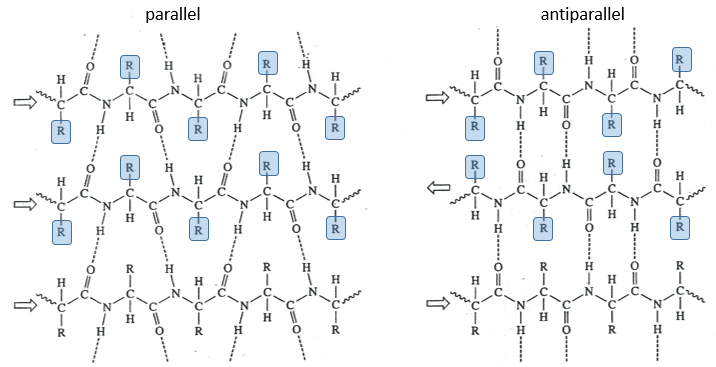
The β sheet structure is a stretched out structure of the peptide chain. The chains place themselves in parallel or in antiparallel to form a compact and rigid structure.
The chains are bond by H bonds and the distance between the chains is 0.35nm. The R groups of the AA have to be small to fit in the space between the chains. Even smaller in the antiparallel β sheets because the R groups are head to head with the R groups of the other chains. In the parallel β sheets, the R groups face one hydrogen. This kind of structure is also largely found in fibrous proteins such as the fibroin (in the silk) or the β-keratin (in feather).
There are also β helices, formed of β sheets arranged in one helix, but these are less frequent.
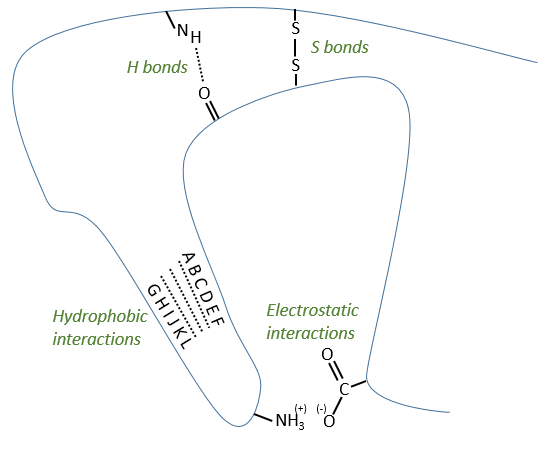
Tertiary structure
The tertiary structure is the result of long distance interactions. In a general way we represent helices by tubes/helices and β structures by arrows. The extremities of the chain are indicated by NH3+ and COO–.
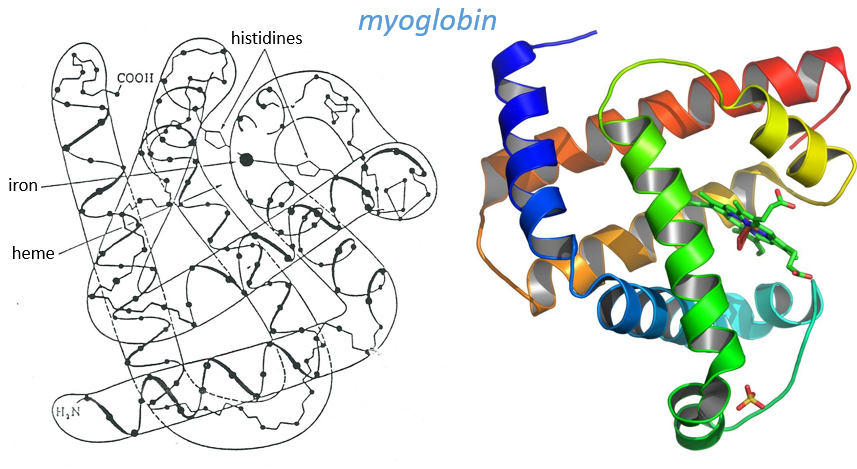
Kendraw determined the tertiary structure of the myoglobin of cachalots. To do so he marked with heavy atoms some known sequences of the molecules.
The structure is compact and possesses 8 helices of variable sizes. The groups inside the protein are nonpolar at the exception of two histidines. The AA in the helices may vary for different species but the angles remain the same.
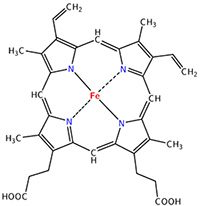
The heme group is the one that fixes the oxygen to supply the muscles. The heme is a tetrapyrol on which one Fe2+ is fixed at the centre.
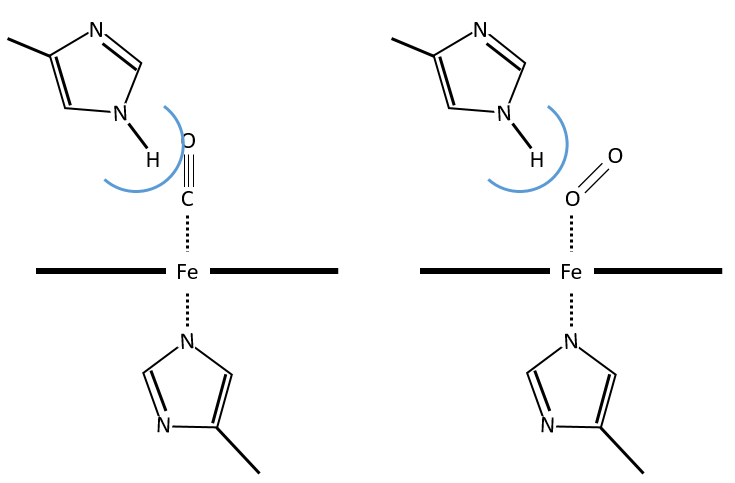
The iron has 6 liaisons and the two last are perpendicular to the plane made by the 4 other liaisons and bind with the two histidine’s. The heme is in the hydrophobic part of the protein because otherwise the iron would be oxidised by the blood. O2 fixes on the iron instead of one histidine. Yet, the presence of the histidine is very important: oxygen can easily bind on the heme but CO has an affinity for the heme 2500 larger than the one of the O2 without the presence of histidine’s. So if there are a few molecules of CO in the air, they would bind to the myoglobin and stay on it for a long time, forbidding oxygen to reach the muscles. The way to heal such an asphyxia is to place the person in a hyperbaric box full of oxygen (and obviously without CO) where we play on the equilibrium to replace the CO by O2. If there are 5000 molecules of O2 for each CO in the air, then the O2 is more susceptible to bind with the haem. Fortunately, histidine’s are close to the iron and prevents sterically the CO to bind correctly with it. The oxygen is not obstructed by the histidine because the angle of liaison is not the same (see the figure below). Considering the histidine, the affinity for the CO drops down to 200 time the one of the oxygen.
Types of liaisons in the tertiary structure:
- hydrophobic interactions
- electrostatic liaisons
- S bonds (cysteines)
- H bonds
Denaturation of the tertiary structure
- by heating: the heat is enough to break weak liaisons such as the hydrophobic interactions. As they break, the structure shambles and the hydrophobic parts enter in contact with the aqueous environment. As a result, the protein precipitates
- by urea: the urea breaks the H bonds
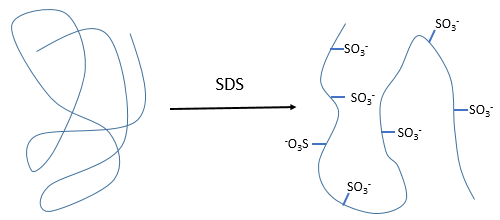
- by detergent: it denatures and solubilises the protein. For instance the SDS (sodium dodecylsulfate).
The hydrophobic chain of the detergent interacts with hydrophobic sequences of proteins. At this point the SO3– is transferred on the protein, generating some hydrophilic zones that modify the structure of the protein.
Quaternary structure
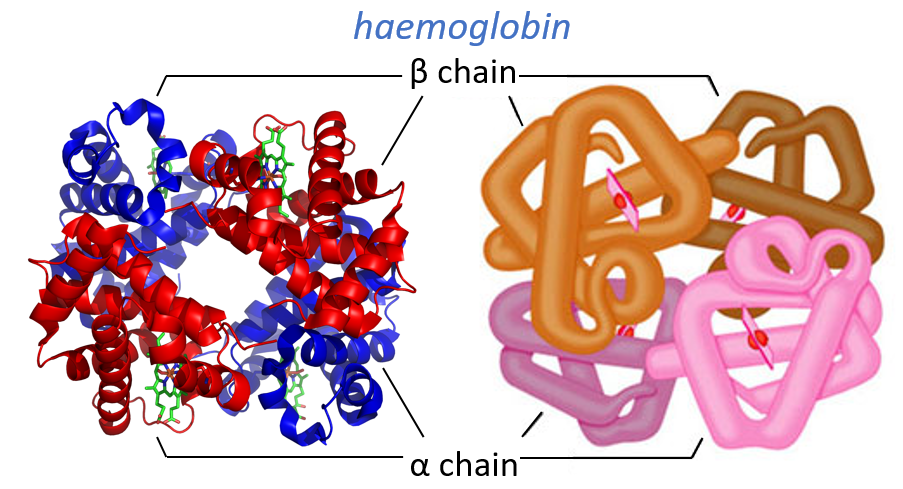
Some proteins aggregate together to form a bigger structure. The parts of this structure are called protomers and are only active if they are together. For instance, the haemoglobin is composed of four protomers, two α and two β.
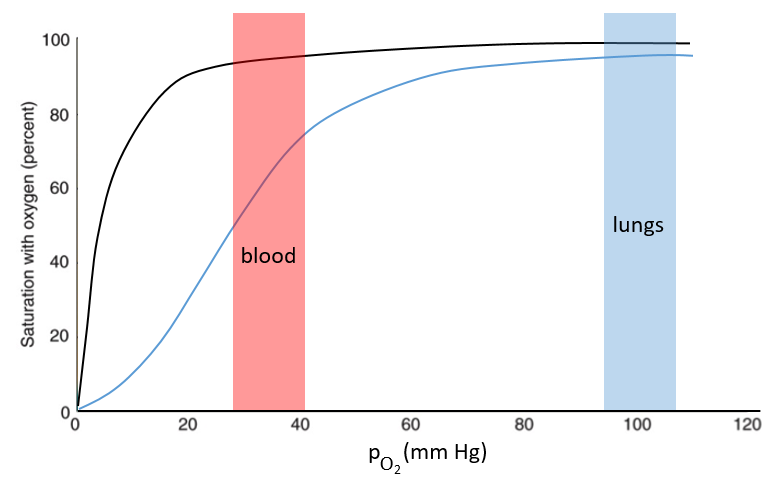
The function of the haemoglobin is to transport the oxygen in the blood and to transfer it to the myoglobin at the muscles. By the way, the protomers are alike with the myoglobin but only in the tertiary structure. The primary structures are very different except at the corners. To give an idea of the role of the haemoglobin, without them there would be 5ml of O2 by litre of blood. With them in the blood, there are 250ml of O2 by litre of blood. Another difference between the myoglobin and the haemoglobin is their curve of fixation of the oxygen. i.e. the percentage of saturation of the protein as a function of the partial pressure in O2.
The curves have similar shapes but the one of the haemoglobin is shifted in larger pressures of O2 In other words, and as expected, the two proteins don’t fix the oxygen at the same place. The partial pressure is high in the lungs and is around 40mm Hg in the muscles and tissues. The two proteins are saturated in oxygen at high pressures but the myoglobin has a rate of saturation that increases much earlier than the one of the haemoglobin. As the myoglobin is only found in the muscles, the haemoglobin fixes oxygen in the lungs and transports it to the muscles. In the muscles, the myoglobin has approximatively 40% more affinity for the oxygen than the haemoglobin. Haemoglobin drops some of the oxygen that is collected to the myoglobin.
The difference of behaviour is explained by the structure of the haemoglobin. The fixation of the first oxygen is difficult and induces a modification of the conformation of the protein through the rotation of two protomers. It opens up the way to a larger central cavity of the protein and the oxygen can now enter more easily. The form without oxygen is called deoxyhaemoglobin and the form with oxygen is called oxyhaemoglobin.
In the cavity, there are some charged amino acids that may enter in interaction with diphosphoglycerate (DPG), a molecule present near muscles and tissues but not in the lungs. With the DPG on the haemoglobin, the curve of fixation of oxygen goes down. By selectively binding to deoxyhaemoglobin, BPG makes it harder for oxygen to bind haemoglobin and more likely to be released to adjacent tissues. The concentration of DPG depends upon the location in the body but we find it essentially in the muscles and in the blood, what explains the difference of the curves of the myoglobin and of the haemoglobin. When we go in altitude, the pressure decreases significantly and we also produce more DPG. It allows the muscles to work better because there is a better transfer of oxygen from the haemoglobin. The DPG is not immediately degraded when we come back to a normal altitude and the muscles continue to be more efficient for a period of time. It is why athletes are often going to the mountain to practice before important events.
The Bohr effect
Haemoglobin can also fix CO2 and protons (both are products of the muscles). Their fixation favours the exchange of oxygen at low pressures. Basically, when we do sport, or run away from a danger, muscles work, produce CO2 and protons. The body uses those products as a signal to ask for more oxygen to maintain the effort. It is an allosteric effect: the activity of the protein is modified by agents that act on a different place of the protein than the active site. The CO2 is fixed on the terminal NH2 of the peptide chains while the protons are fixed on histidines.
Enzymes
Enzymes catalyse chemical reactions with a huge specificity. Usually there is only one target substrate but sometimes other molecules can take place in the active site and block it. The names of the enzymes are often related to the target substrate. For instance, the enzymes responsible for the cleavage of RNA is the RNAase, of lipids a lipidase, of amino acids a protease.
The activity of enzymes is modulated by several factors
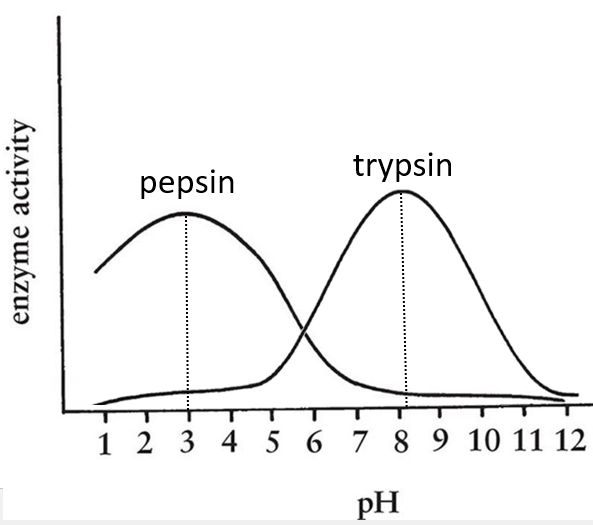
- pH: there is usually a maximum of activity at a given pH. For instance the pepsin and the trypsin are two enzymes that are respectively active in the stomach and in the intestine. It is thus normal that the optimum of pH of the pepsin is much lower (more acidic, pH»3) than the one of the trypsin (pH»8).
- cofactors: they can be ions or coenzymes and their presence will affect the activity of the enzyme. As for the pH, there can be a maximum of activity depending on the concentration of the cofactor.
- temperature: chemical reactions obey to the law of Arrhenius
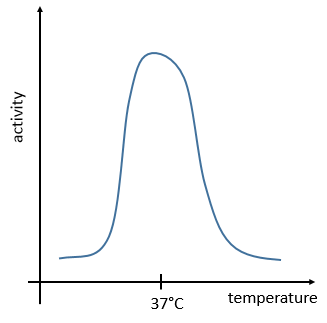
The temperature increases the speed of reaction but if it increases too much the enzyme is denatured. There is thus also an optimum of temperature. In the body, this optimum is usually around 37°C.
- saturation: the concentration of enzyme is often small in comparison to the concentration of substrate so it limits the maximum speed of reaction. If we increase the concentration of enzyme, the maximum speed increases but it is only true up to a given point. After this point, we can increase the concentration of enzyme but the speed will not change. It is translated by the constant of Michaelis-Menten (see the chapter on kinetics).
A small KM means that the enzyme reaches its full speed for lower concentrations. Half of the maximum speed is reached when the concentration of substrate equals KM.
Chapter 4 : Primary structure of proteins
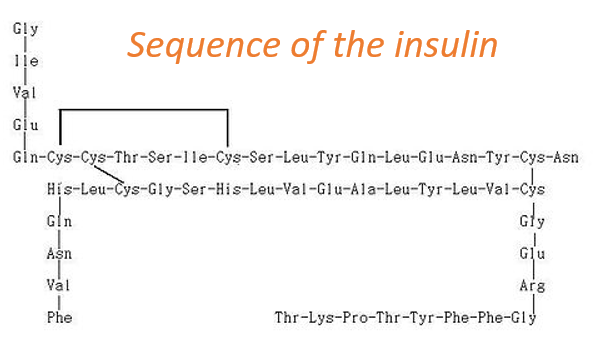
The primary structure of a protein is the succession, or sequence, of the AA. Proteins are made of one linear chain of amino acids. This linear chain takes a 3D structure (secondary and tertiary structures) because of the interactions between the sequences in the chain and the interactions (hydrophobic or hydrophilic) with the surrounding. The order of the AA and their quantities are well defined for each protein. The first structure ever determined was the one of the insulin. This protein is 51 AA long, what is small in comparison with other proteins that can be up to 8000 AA long.
At the exception of the disulphide bridges between the cysteines, this protein may be seen as two linear chains with respectively 21 and 30 AA. As the insulin is not composed of one single chain, this protein is obviously not the primary product of a ribosome. Some further operations were applied to the precursor forms of the insulin to obtain the functional protein. We will see this process in more details later.
To determine the primary structure of a protein, we need it to be pure. To separate the proteins from the other molecules, we perform a dialyse: the solution containing the proteins in a bag with a porous membrane through which small ions can pass but not macromolecules. Once the equilibrium is reached, we repeat the process to obtain the best purity possible. We still have to separate macromolecules. It is done
- by chromatography
- ionic: the proteins have a charge that depends on the R of the amino acids. The charge depends on the solvent that is used. At one given pH, the target protein is positively charged. Placing a polymer negatively charged in the column will retain the protein while the others pass through the column. After a while we change of solvent to unbind the protein from the polymer.
- of exclusion: the proteins are separated as a function of their size. The large protein move faster than the small ones because they cannot enter in the holes of the beads composing the immobile phase.
- ligands: the protein can bind with specific ligands. Normally only this protein should bind, what assures a great purity.
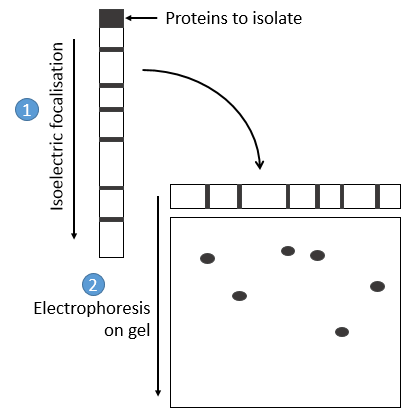
- by electrophoresis: the proteins are separated on the basis of their isoelectric point: the proteins are placed on a gel of polyaccrylamine the pH of which is not uniform but a gradient. As the proteins are charged in function of the pH of the solvent, it will move until its charge becomes neutral. After that, we take the gel and apply it on top of a second, long porous gel where the proteins are displaced by the application of a charge and their speed also depends on the size of the protein. This way we obtain a 2D separation of the species.
Once the proteins are isolated, we can start to determine its structure. To do so, we use an exopeptidase: its role is to cleave only the liaison before the last AA. The problem is that the enzymes are not synchronic: they don’t all start simultaneously and some chains may be cleaved several times while others are cleaved once or not at all. There can thus be a mix of amino acids in the solution and the determination of the right sequence may be difficult. To minimize this problem, the experiment has to be very short.
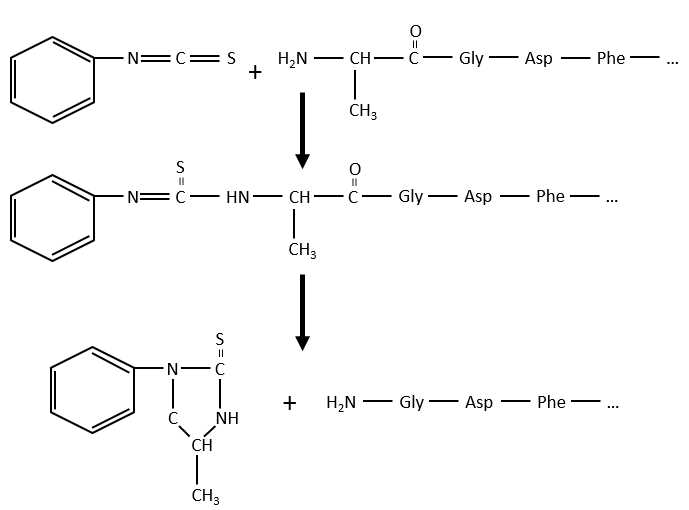
Another method (of Edman) is to weaken the last peptide bond and next to use a weak reactant that hydrolyses only this liaison. In the presented case, a phenyl isothiocyanate reacts with the amino-terminal residue to form a thioamide. The next peptide bond is weakened and can be cleaved without hydrolysis because of the formation of a cycle.
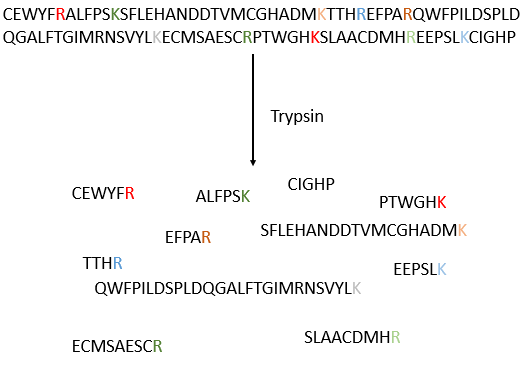
The process can be repeated several times to determine the initial amino-terminal sequence but the yield of the reaction is not of 100%. As a result, it can only be repeated around 30 times. Moreover, sometimes chains break and we have thus a new extremity that gives different results. Anyway 30 AA is not good enough to determine the structure of proteins that can be 8000 AA long. The order of the 30 first amino acids don’t give any clue on the nature of the following amino acids. The only way to determine the structure is to first fragmentise the protein into small bits. We will try to cleave the protein only between some amino acids to have points of reference. There are some specific endopeptidases enzymes that cleave proteins only after one particular amino acid. It detect the presence of one specific R, binds to it and cleaves the peptide bond. The trypsin is one enzyme that cleaves the peptide bond after one lysine or arginine.
We separate the fragments by chromatography and determine the sequence of each fragment (with the method of Edman for instance). Yet, we don’t know in which order the fragments are in the protein. The process is repeated with another specific endopeptidase. After several fragmentations and sequencings, we can try to figure out which sequences are in common in different fragmentations and reconstruct the whole sequence.
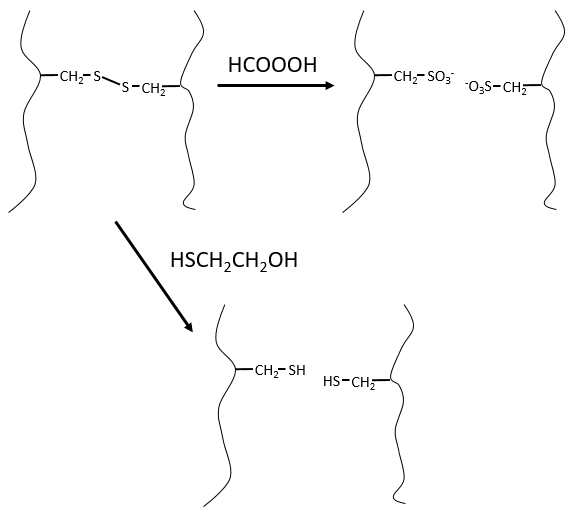
It would be perfect if there were no SS bonds along the peptide chain. There is thus a step that has to be done prior to the fragmentation: we use performic acid (HCOOOH) to oxidise the sulphurs. This method is irreversible. A second method, reversible, can be used instead: the mercaptoethanol reduces the sulphurs.
We can resume the primary structure in 4 points:
- the chain is linear and non-branched.
- between the amino acids, the only covalent bonds are the peptide bonds and the SS bonds between the cysteines. The other interactions are attraction/repulsion between charged groups and H bonds.
- the sequence is completely defined and arbitrary: one part of the sequence does not defines the rest of the sequence. For instance, if we know 10 amino acids, we cannot say that the 5 next are a guanine, a cysteine, etc…
- proteins from different species are similar but not identical. There are some key fragments that are common but the whole structure may vary. One exception is the histone. This protein is common to all the eukaryote species: it is the protein responsible for the compaction of the DNA in the cellular nucleus. Even if the information contained in the DNA differs between species, its structure is identical and the protein responsible for its coiling/uncoiling is thus the same for all the eukaryote species.
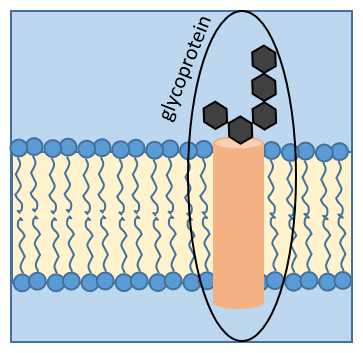
Some proteins fix one (or more) monosaccharide. This process is called glycosylation and lead to one glycoprotein.
The monosaccharide can be fixed on one nitrogen atom (N-linked glycosylation, exclusively on the asparagine) or one oxygen atom (O-linked glycosylation). The monosaccharide can itself be bound to a chain of monosaccharides. We frequently observe glycoproteins in membranes where they play the role of receptors. The presence of the monosaccharide on the protein permits a great diversity in the receptors for the cells.
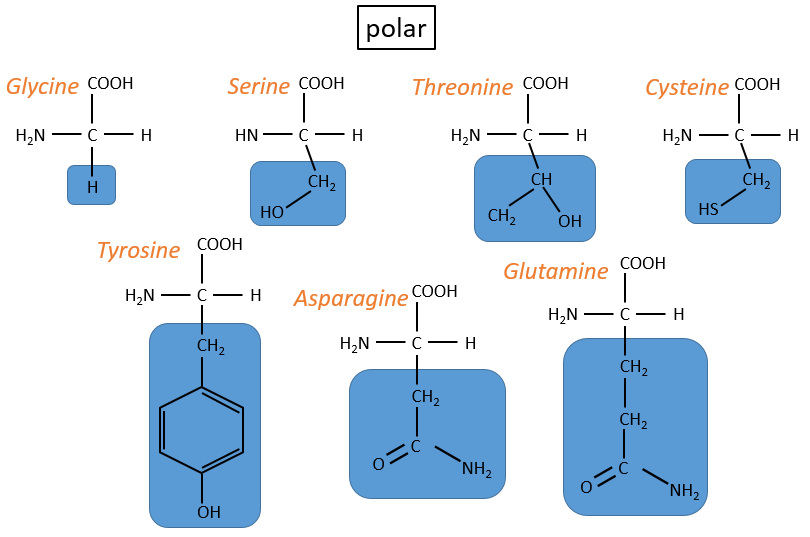
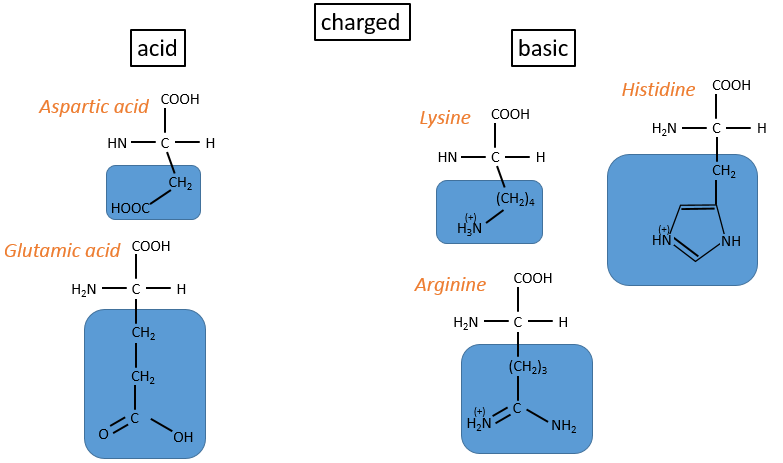
Chapter 3 : Proteins and amino acids
The word protein comes from the Greek word proteos, which means first. Proteins are indeed one essential element of the life. They are well defined in composition, size and shape and each one has a very precise role: transport, defence, hormones, … Some have also an exotic role. For instance some proteins prevent the blood of fish to freeze. We can however sort the proteins in three main classes: fibrous, globular and membrane proteins. The fibrous proteins are stretched out and fragile. Two examples are the keratin and the collagen. The keratin is one tough and insoluble fibrous structural proteins that composes hair, wool, horns, nails, claws, etc. The toughness of the keratin is rivalled only by the chitin in biological materials. They are not only localised in the hard parts of animals but are found in all epithelial cells, i.e. cells that cover the external surfaces of organisms and internal surfaces of organs to reinforce their structure. Horns, claws, nails … are produced by epithelial cells adapted to growing an abundance of keratin and then dying as individual cells while leaving the keratin to help form a structure valuable to the whole animal. The collagen is the main structural protein in the extracellular space, making it the most abundant protein in mammals. They form bones, tendons, cartilage in function of the local degree of mineralization. Membrane proteins are found in membranes and serve as receptors or provide channels to transport ions or molecules from one side of the membrane to the other side. Globular protein are basically the non-fibrous proteins and have diverse functions (example: haemoglobin, lysozyme). All the enzymes are globular proteins.
One common point between all the proteins is that they are insoluble in organic solvents. Thanks to that, we were able to determine their (crystalline) structure and composition. For instance, in the bones we found the collagen by crystallisation. We can destroy the protein to determine the amino acids it is made of: glycol. In the silk, we discovered the fibroin, composed of serine.
Amino acids (AA)
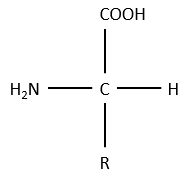
The general structure of amino acids is
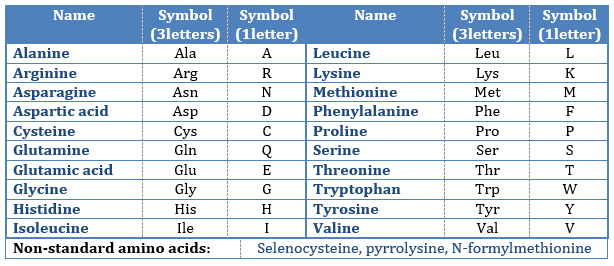
where R is the group that makes the distinction between all the different amino acids (with one exception) .They are zwitterions: they wear an acid group (COOH) and a basic group (NH2). The central carbon is chiral and the AA can be L or D. 20 AA are found in the proteins (over 23 AA).
The AA are usually written by an abbreviation of 3 letters are sorted in function of their functional group R
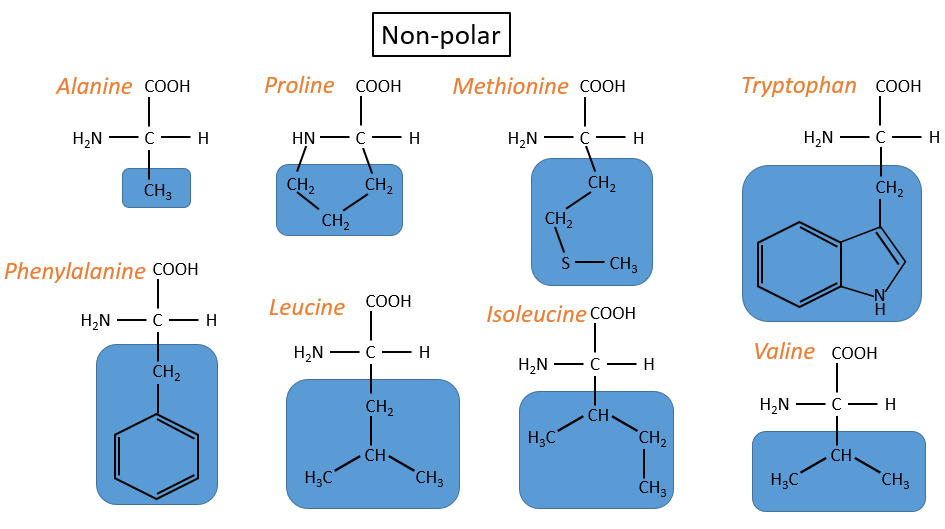
- non-polar hydrophobic: no interaction with water
- proline: its R is cyclic and binds to the amine group of the AA. It forbids the rotation of the AA.
- methionine: it possesses a sulphur atom
- neutral polar: may react with the solvent. They are polar because R is very small
- cysteine: they bind together via S bonds
- charged polar
- acids: with COOH
- basic: with amine derivatives
The isoleucine and the threonine have one asymmetric carbon. Only the L are natural.
As zwitterions, the amino acids have an isoelectric point that depends on the nature of R. We can easily determine the composition of one protein thanks to this property: once hydrolysed, the amino acids that compose the protein can be separated by chromatography or electrophoresis to determine their proportions. There is also a way to determine the last amino acid of the protein (ended by NH2): prior to hydrolysis, we use the reactant of Sanger (1-fluoro-2-4-dinitrobenzene). This coloured group is fixed on the amine and the bond cannot be hydrolysed. We can thus identify it after the chromatography. Another method is the degradation of Edman. In this case a phenyl isothiocyanate reacts with the amino-terminal residue to form a thioamide. The next peptide bond is weakened and can be cleaved without hydrolysis because of the formation of a cycle.
The process can be repeated several times to determine the initial amino-terminal sequence.
The interactions between the AA of the protein with the other AA and with the solvent are responsible for the structure of the protein.
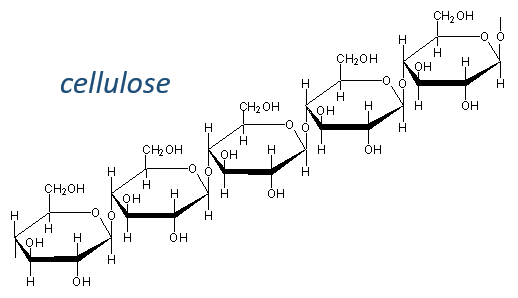
Chapter 2 : Glucides
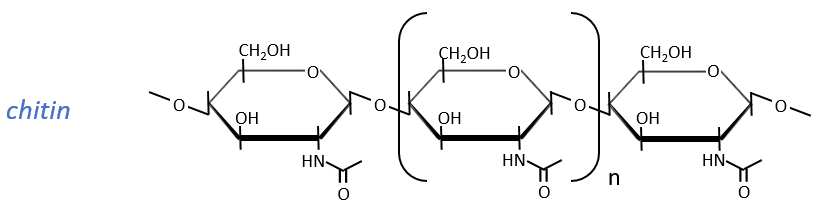
Glucides are essentially cyclic hydrates of carbon (CN(H2O)N) but can also wear N, S, P. The cycles are usually 5 or 6 atoms long and form macromolecules when cycles bind together. One example is the cellulose, which is the walls of vegetables. As humans, we don’t properly digest the cellulose contained in the salads we eat so we have to masticate it more to break it. Another example is the chitin which is the essential component of the shell of insects, crabs, etc.
The nomenclature of the monocycles (or monomers in we consider the macromolecules as polymers) is to end their name by ~monosaccharide. The most known are the glucose and the fructose, which differ by the presence of one ketone in the fructose. The glucose is the most abundant monosaccharide and represents 50% of the carbons on Earth. Algae’s produce billions of monosaccharides by year and require the sun light to do so. In the industry, we use monosaccharides in paper, cotton, drinks, food, pharmaceutic products, …
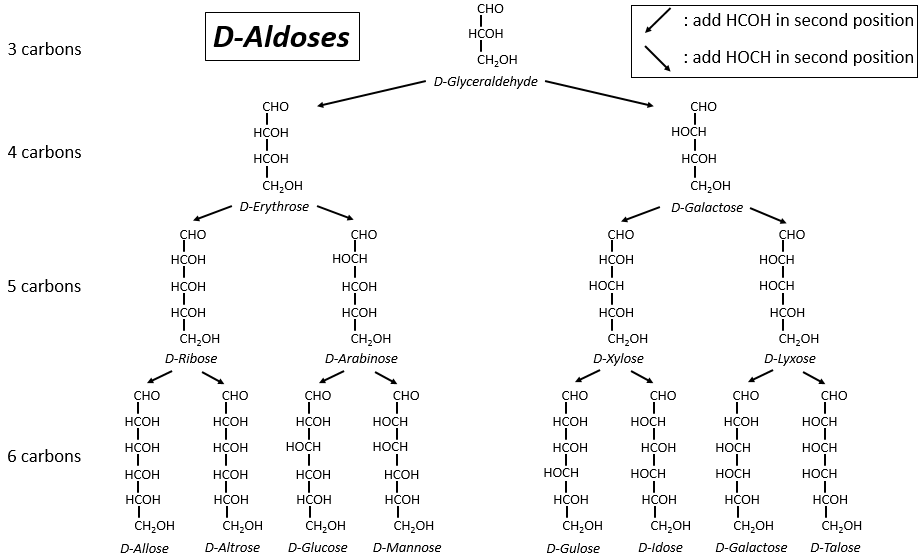
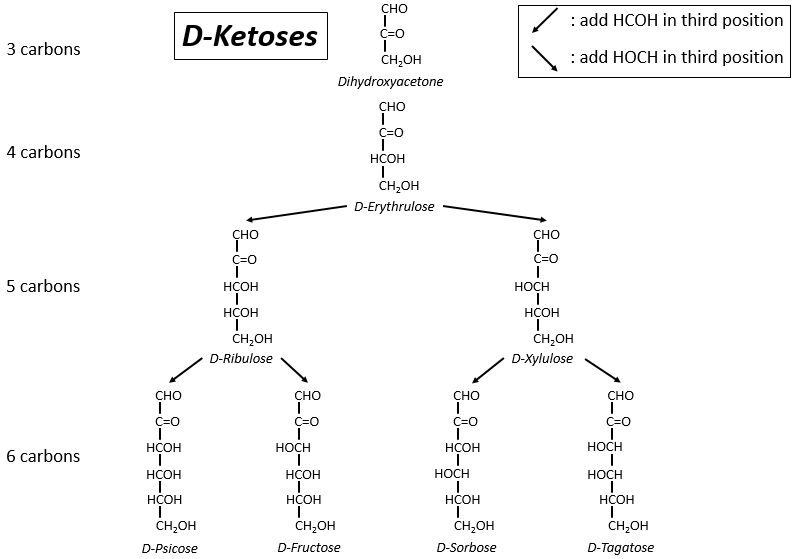
The following tables show the projections of Fischer of sugars. Basically, the chains are composed of carbons wearing –OH functions. At the top extremity there is an aldehyde in the case of aldoses and a ketone in the case of ketoses. It is the first parameter of sorting. The second parameter is the length of the chain, going from 3 to 8 (chains of 8 carbons are only artificially made).
One may have seen that the –OH are not all on the same side of the chain. The carbons inside the chain (not the ones at the extremities) are chiral and the sugars have thus an optical activity. The tables we showed previously only show the dextrogyre molecules, noted with the prefix D-, i.e. the ones that deflect the light towards the right. The distinction is not only important for the light: most of the enzymes accept only one of the conformers, L (levogyre) or D (dextrogyre).
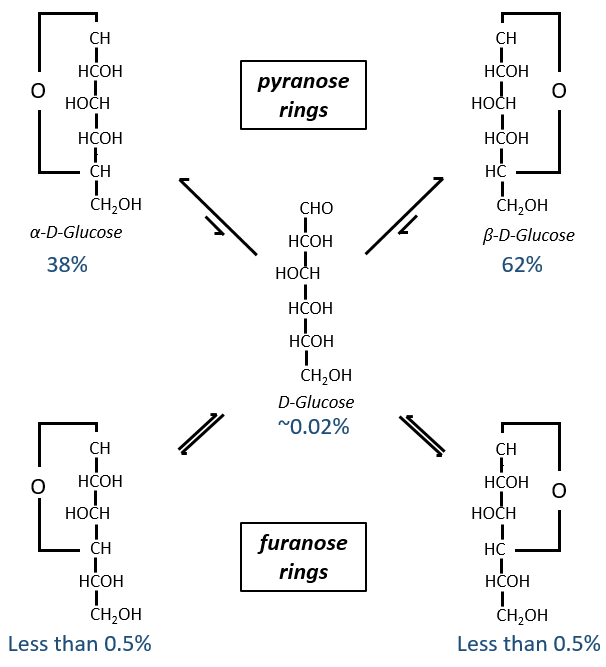
Now you will tell me that I began this section saying that the glucides are cyclic and that I only showed linear molecules. In aqueous conditions, the aldehyde and one hydroxyl function (in position 4 or 5) merge to form one hemiacetal, a cycle of 5 (furanose rings) or 6 atoms (pyranose rings).
The equilibrium of this process is almost towards the cyclic forms with 6 atoms, i.e. the pyranose rings. The crystals of the linear sugars were obtained in pyridine in which the rotatory power is +19° for the glucose and in the water it was +53°.
This rotatory power is the averaged value of the species in the water (the α-D-glucose has [α]D=120 and the β-D-glucose has [α]D=19).
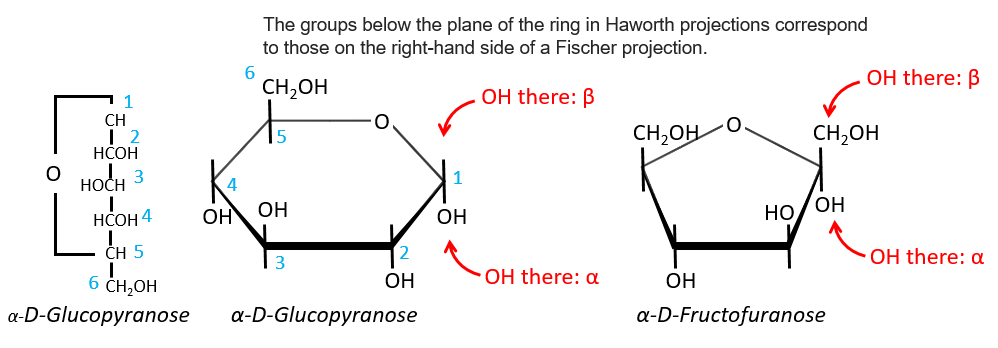
Projection of Haworth
The cycle is represented flat. The heterocyclic oxygen is on the top right position and the chain outside of the cycle is on top left position of the cycle. The –OH groups are placed on top of the cycle if they are on the left in the projection of Fischer and under the cycle if they are on the right of the chain in the projection of Fischer.
We enumerate the carbons starting from the right and moving in the clockwise direction. The C1 is the highest carbon of the Fischer projection and is here an anomeric carbon: when the glucose was put in water, the OH group could be on the left or right of the chain. Its position determines if the cycle is α or β and we can pass from the α to the β forms only by the linear form. We have the same kind of representation for furanose rings and the cyclisation is also done in the case of ketoses and of pentoses. Also note that the C1 carbon may be out of the cycle, as it is the case for the α-D-fructofuranose shown above.
Osides are monosaccharides bound to one molecule on the anomeric spot. The presence of this molecule there blocks the configuration of the monosaccharide: it cannot change from α to β or change for its linear form anymore. As the anomeric carbon is not available, the reduction power of the monosaccharide is also lost.
Disaccharides
They are combinations of two monosaccharides. We will use them to show where the liaisons can be and how we represent those molecules in the representation of Haworth.
- Maltose
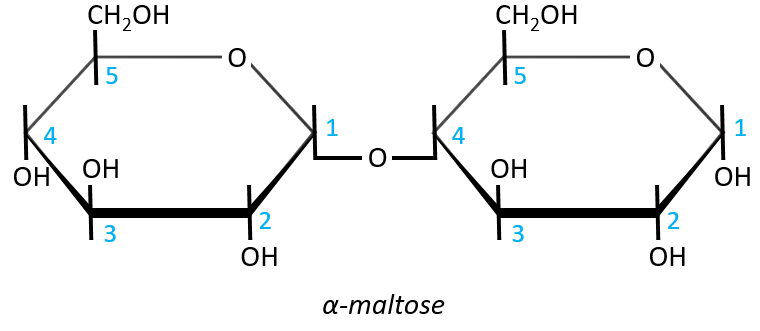
It is the combination of two glucoses bound together in (1→4).
The liaison is made between the C1 of the glucose of the left and the C4 of the glucose of the right, indicated by (1→4). We let the representation of both glucoses as they were for the monosaccharides alone and bind them with a straight line. It is as simple as that. One anomeric carbon is taken by the liaison but the second is free. The molecule is thus a reductant and able to do the mutarotation.
The nomenclature of the α-maltose is
α-D-Glucopyranosyl-(1→4)-D-glucose
or also
4-O-α-D-Glucopyranosyl-D-glucose
The monosaccharide with its anomeric carbon free ends the name while the other monosaccharide name is ended by ~osyl. The position of the liaison between the monosaccharides is indicated by the (1→4) written between the monosaccharides or the 4-O prefix: liaisons between monosaccharides are always involving C1 so we just indicate the second carbon involved in the liaison (C4) and the fact that the liaison involves and oxygen atom. The alpha also gives an information on the liaison between the sugars: the type of the anomeric carbon that is now bound to the second monosaccharide. The dextro or levogyre character of each monosaccharide is indicated as well.
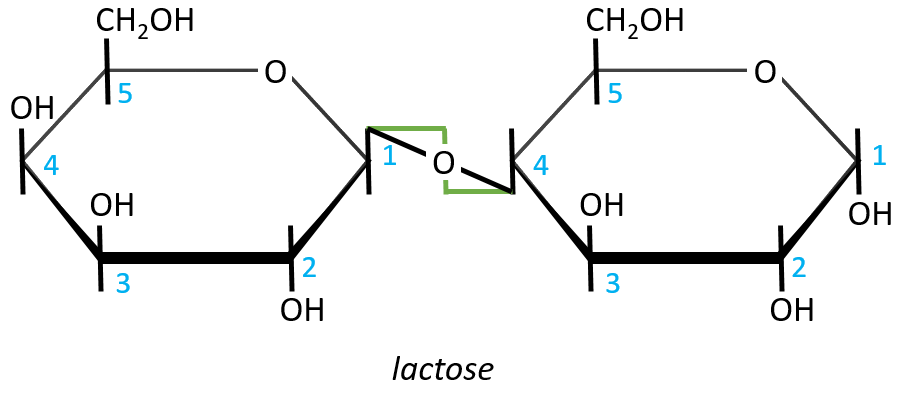
- Lactose
It is the combination of one β-galactose with one α-glucose. The liaison is also in (1→4).
In this representation, we maintain both representations of the monosaccharides. The liaison between them is thus represented as a diagonal or as a S shaped line as showed above in green. The nomenclature is
β-D-galactopyranosyl-(1→4)-D-glucose
or
4-O-β-D-galactopyranosyl-D-glucose
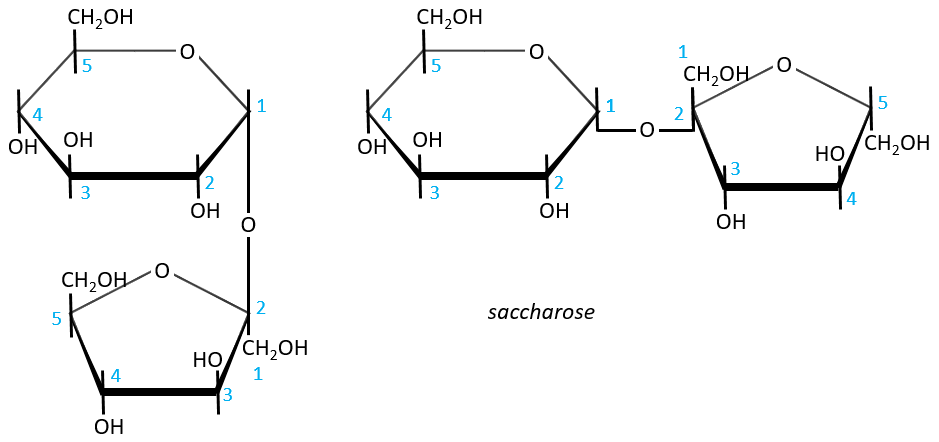
- Sucrose (or saccharose)
It is an example of disaccharide for which the liaison is not 1à4. The saccharose is the combination of a α-glucose with a β-fructose. They are bond by their anomeric carbons, i.e. in (1→2).
There is thus no possibility of mutarotation. The nomenclature is α-D-glucopyranosyl-(1→2)-β-D-fructofuranoside. The name doesn’t end by ose because the anomeric carbon is not free.
The invertase is an enzyme that is able to cleave the liaison between those two monosaccharides. This enzyme is interesting for the industry because the saccharose has a sweetening power way smaller than the sum of the sweetening powers of the fructose and of the glucose.
Polysaccharides
They are chains of monosaccharides. We can sort them in two types: homopolyosides that are chains of one single monosaccharide that is repeated, and heteropolyosides that are (usually) two different monosaccharides that are repeated with a given order or at random. The size of the chain can be very long (thousands to hundreds of thousands of monosaccharides).
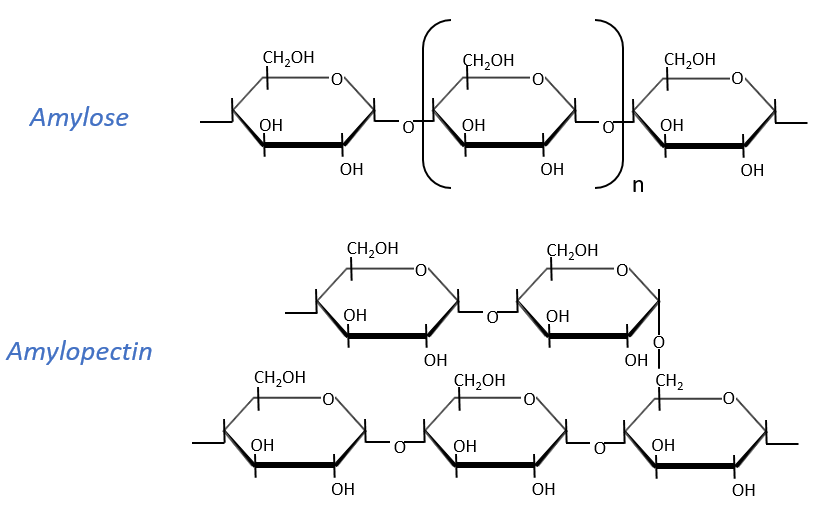
Starch is one macromolecule we find in vegetables, composed of monosaccharides. There are separated in two groups: the amylose (15-20%) and the amylopectin (80-85%). The amylose is a chain of glucoses linked in (1→4) 100 to 300 units long while the amylopectin has ramifications every 30 sugars approximatively, linked in (1→6). In animals, the equivalent of the amylopectin is the glycogen for which the ramifications are more frequent (every 8 to 12 sugar). The amylose forms a α helix (6 to 8 glucoses form one whorl).
Enzymes can cleave liaisons to free glucose molecules. Some enzymes cleave in the chains (endoglycosidases), some at their extremities (exoglycosidases). Given the size of the starch and the amount of extremities, several enzymes can act simultaneously.
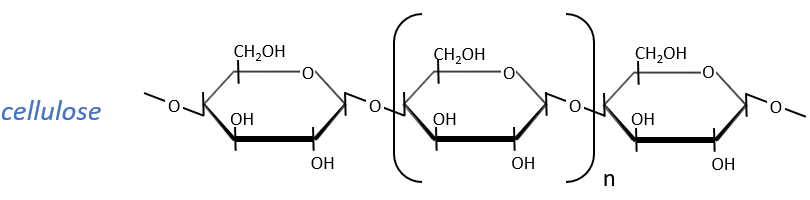
The cellulose is made of β-glucoses, up to 10000 units. This difference with the amylose leads to a huge difference of conformation: instead of helixes, the cellulose forms planes bound together by H bonds. Moreover, the β liaisons cannot be cleaved by the enzymes of most of the animals. Only ruminants are able to do it because they have cellulases, enzymes able to cleave the cellulose.
The chitin is similar to the cellulose except that the beta glucoses are replaced by a variant (N-acetylglucosamine). Combined to Ca2CO3 it forms exoskeletons of insects.
Heteropolyosides
Glycosaminoglycan
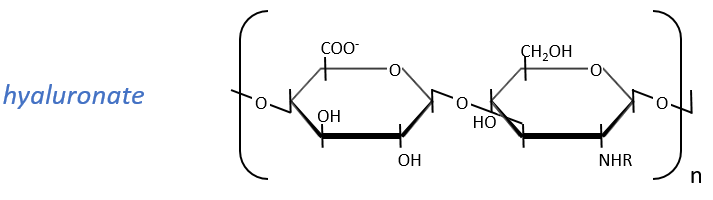
They are found at the outside of the cells, bound to the lipid bilayer and allow the movements of the cell (one motor is in the lipid bilayer). An example is the hyaluronic acid (the cosmetics industrials highlight it nowadays).
This molecule is a polymer composed of a repetition of a block of two monosaccharides, the β-glucoronate and one N-acetylglucosamine. They are bound in (1→4) and in (1→3) to form one chain. As the β-glucoronate is negatively charged, the chains don’t interfere one with each other. Chains are bound to proteins
Peptidoglycans
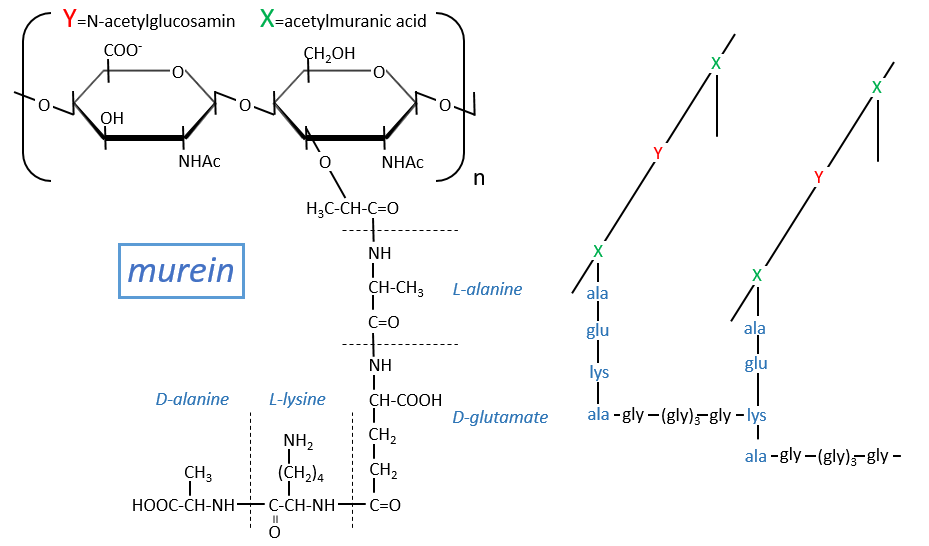
Some monosaccharides of the chain wear a peptide chain of amino acids. The peptide chains bind together to form a network with a given rigidity. Peptidoglycans are also known as the murein.
It is the rigid constituent of the membrane of bacteria’s. It is composed of a block of N-acetylglucosyl (a derivative of the glucose) bound in (1→4) with the N-acetylmuranic acid, the same monosaccharide but wearing one peptide chain on C3. The chain is bound to another chain by a liaison between several glycine’s and lysine to form a dense network. The peptides in the chains are consecutively L, D, L, D, …
The mechanism of some antibiotics is to break the liaison between the monosaccharides to destroy the whole structure.
Chapter 1: Biochemistry – Introduction and lipids
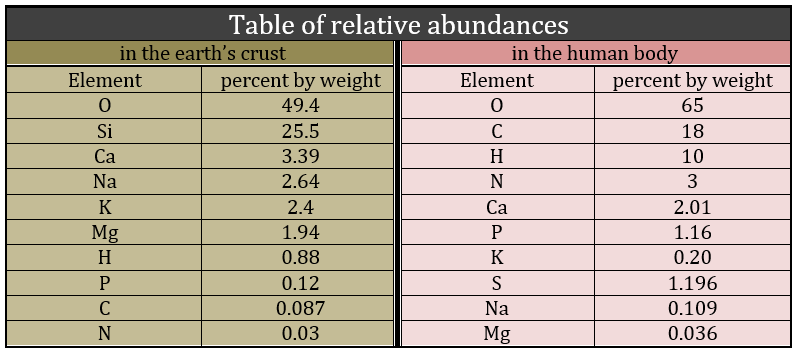
Biochemistry is a field of the chemistry related to the living bodies, animal or vegetal. This field is far from the inorganic chemistry and a simple comparison can show it: the repartition of the molecules in the earth’s crust and in a living body are totally different. The first is essentially composed of silica and the second of carbon.
Historically, the first theory about the life was that it is given by God and only by Him. The molecules are inanimate and cannot be turned into a living body except if God breathe life into it. It is called the vitalism theory.
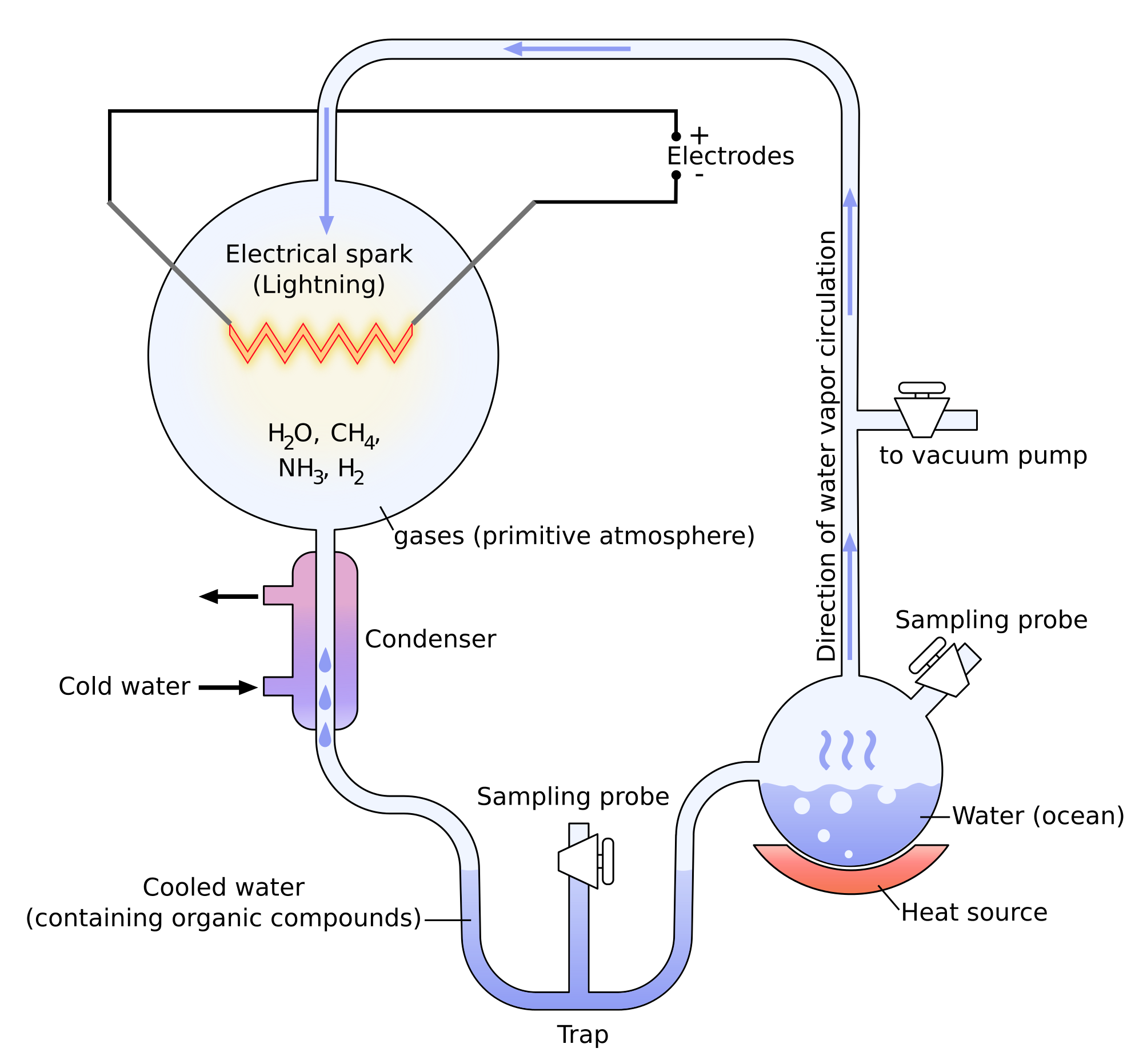
Miller showed through its experiments that it was possible to create molecules belonging to living bodies with simple molecules. The experiments of Miller consist in the modelling of the conditions on Earth before the apparition of the life. There was not a lot of oxygen at this time and the atmosphere was essentially composed of ammoniac, methane, hydrogen and water. The temperature was way higher and there was a lot of lightning. Using these conditions, some molecules that are found in living bodies were spontaneously formed: urea, glycine, alanine, …
Yet there was no trace of proteins and especially of RNA (ribonucleic acids), the molecules capable to write DNA (deoxyribonucleic acids) molecules. It was showed later that some molecules of meteorites allowed the formation of RNA.
Principal characteristics of biochemistry
Limitations
Amongst the good hundreds of atoms in the table of Mendeleev, we only find 16 different liaisons in biochemistry but it allows the formation of an infinity of organic molecules. That is even more surprising that the reactions involved in the biological processes are limited by the biological conditions of existence: there is no way that a reaction that requires a temperature of 100°C takes place in our body. It would damage the surrounding molecules and burn tissues. As any chemical reaction, they must obey the rules of thermodynamic and find the energy required to make the reaction somewhere else.
Specialisation
In a body, the reactions are made in cells and can also be compartmentalised inside the cell. All the cells cannot do all the reactions and some cells are specialised. The specialisation is written in the genetic information and is translated by the presence or the absence of some enzymes. Moreover, in one cell there are not the same kind of reactions in the mitochondria than in the endoplasmic reticulum. It is because the reactions are regulated, positively or negatively, by catalysts.
Enzymes
The temperature has to remain low (~37°C) and cannot vary a lot. ∆G=∆H-T∆S has to be negative and close to zero. The speed of reaction would be close to zero without catalysts.
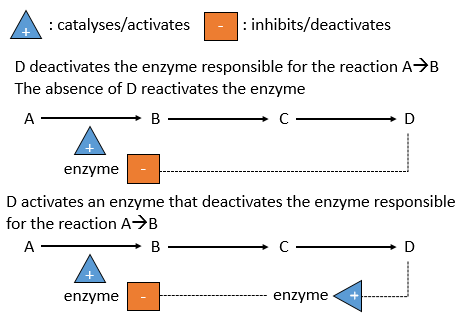
There is a system of regulation in the cells: the activity of the catalysts depends on the requirements of the cell. If the cell is full of one species D, there is no need to accumulate more of it while the resources could be used in a different process. The presence of the species D will influence a catalyst (often an enzyme) that acts on one reaction leading to the formation of D. The presence of D can inhibit an enzyme responsible of the reaction or activate an enzyme inhibiting the reaction. On the contrary, the absence of D can also be detected by this enzyme or another to favour the formation of D.
The management of the energy is a global problem for the body: the processes cannot consume or reject too much heat at once. It could damage the surrounding cells and inhibit most of the enzymes: the temperature of the body is usually in the optimum range of efficiency for the enzymes and if there is a variation of temperature, the activity of the enzymes lowers significantly.
Regulation
Moreover, there are periods during which we consume less energy (when we sleep for instance) and periods of times when the body needs more resources (when doing sport, during a stress, …). These periods do not coincide with the periods during which we produce the energy (when we eat). There must thus be an effective way to store the energy when it is produced and to liberate it when needed. It is done through the formation of ATP (adenosine triphosphate) from ADP (adenosine diphosphate). This reaction is only possible in the mitochondria but the energy is needed everywhere. The energy has thus also to be transported in the different compartments of the cells and in the body.
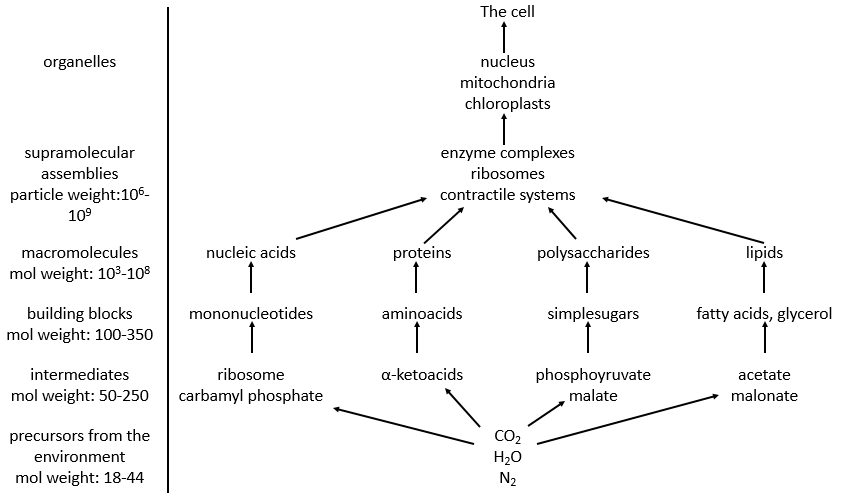
This course will focus on these problems through the example of the degradation of the glucose. Prior to that, we will make an introduction of the main types of molecules that we find the body.
Molecules of the body
This course will focus on these problems through the example of the degradation of the glucose. Prior to that, we will make an introduction of the main types of molecules that we find the body.
Water
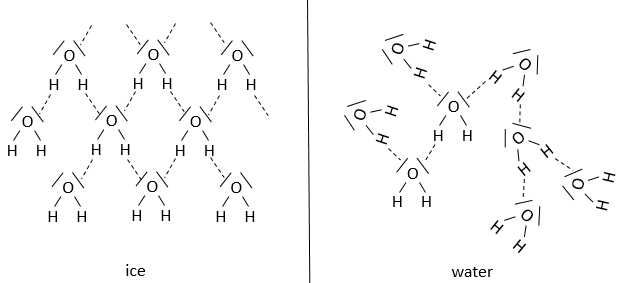
Our body is composed at 60-70% of water. Most of the reactions are thus made in aqueous and polar conditions. The water implies an organisation between the molecules. In the ice, one molecule of water makes 4 H bonds with other H2O molecules.
At the temperature of the body, this number drops to approximatively 3.4. The H bonds break easily and frequently: their energy is ~4.5kcal/mol (against ~ 110kcal/mol for a covalent liaison) and their life time is about 10-9s.
The H bonds bind water molecules together but also water molecule to any electronegative atom (for instance R-OH, R-CO-R, R-COOH, R-NH2…). The groups that form H bonds are called hydrophilic groups. Groups that don’t allow the formation of such bonds are called hydrophobic groups (an aliphatic chain for instance, a phenyl …). The firsts are soluble in water while the seconds are not (they form two separated phases). One molecule that wears hydrophilic and hydrophobic groups is called amphipathic.
The transpiration is a way that the body has find to regulate the temperature: the energy of vaporisation of the water is 540cal/g of water. To compensate the rise of temperature of 1 degree, 2 grams of water are evaporated.
Lipids
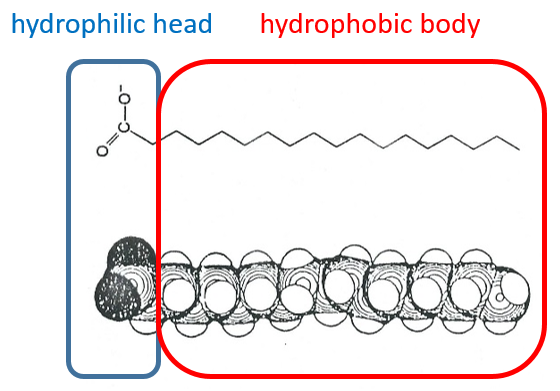
Lipids are a group of naturally occurring molecules usually composed of one hydrophilic head and of one or more hydrophobic chains.
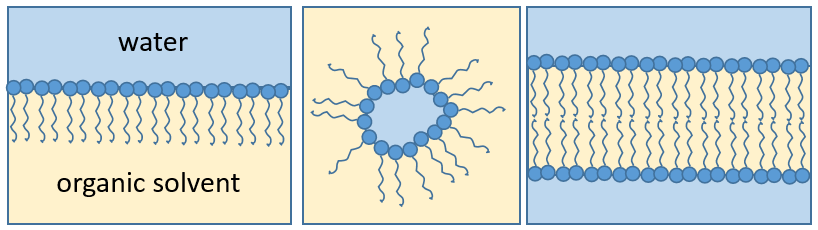
The presence of the polar head does not mean that the whole molecule is soluble in the water. The hydrophobic chains are insoluble in water and aggregate together to minimise the surface of contact with the water molecules. The main biological functions of lipids include storing energy, signalling, and acting as structural components of cell membranes. The membranes they form can either be a monolayer (separating an aqueous and an organic phase) or a bilayer (usually separating two aqueous phases).
The lipids are sorted in function of the ability to form soap. To do so, the lipid has to contain a fatty acid (one hydrophobic chain finishing with a hydrophilic COOH group).
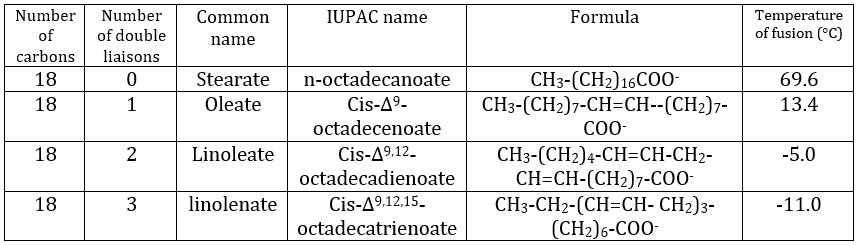
Their nomenclature is ended by ~(an)oate. For instance, CH3(CH2)10COO– (12 carbons) is called dodecanoate. Some lipids wear names coming from the vegetal they come from. For instance CH3(CH2)14COO– is called palmitate because it comes from palm trees. If there is a double liaison in the chain, its position is indicated at the beginning of the name by a ∆x where x is the position of the first carbon with a C=C and the suffix is now ~enoate. For instance, CH3(CH2)CH=CH(CH2)7COO– is called ∆9-hexadecenoate.
The presence of one double liaison hardens the structure of the lipid. One lipid without C=C is said saturated and all the liaisons can rotate. These lipids are very flexible. On the other hand, the C=C fix the conformation locally because the double liaison cannot rotate. As a result, the cohesion between the chains is more complex and the number of interactions of van der Waals decreases (in other words, the chains cannot imbricate as well as saturated chains). A lipid with one double liaison is called unsaturated. If there are several C=C, it is called polyunsaturated. To illustrate the difference one C=C makes, the following table shows a series of lipids with their temperature of fusion.
The unsaturated lipids are found under the form of an oil while the saturated ones under the form of fats. It is possible to hydrogenate the unsaturated lipids to remove the C=C.
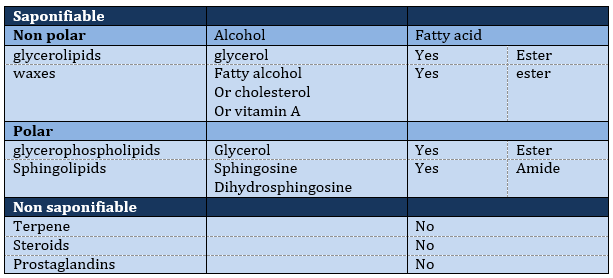
Saponifiable lipids
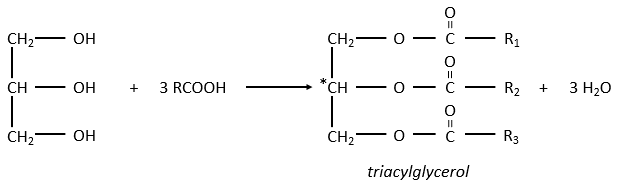
The glycerol is a polyalcohol with 3 –OH groups on a 3 carbons chain. When it reacts with 3 equivalents of RCOO– (a lipid), it forms a triacylglycerol. The carbon noted *C is chiral if R1 and R2 are different. The reaction in the opposite direction is the reaction of saponification:
The reaction in the opposite direction is the reaction of saponification:
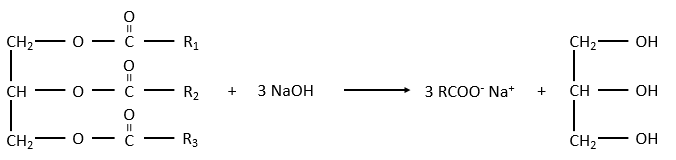
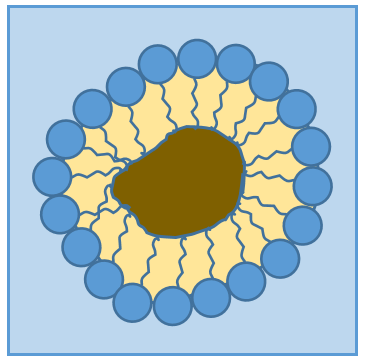
RCOONa is a soap. The principle is quite simple: the soap forms micelles around dirt that are not soluble in water. The hydrophobic chains encircle the dirt and the hydrophilic heads are in contact with the water.
The whole thing is soluble in the water. Lipids are an energetic reservoir stocked under the form of pellets of fat in cells called adipocytes.
Waxes are a combination of a fatty acid with a fatty alcohol.
They are very hydrophobic and are an efficient protection against the humidity (example: the wool of sheep) or to retain the water from evaporating (ex: tropical plants, cactuses). Toothed whales use a mixing of waxes and of triacylglycerol’s, called spermaceti to swim effortless in depths. At 37°C, the mix is an oil but deep the temperature drops and the mix crystallises. Its density increases, what allows the cachalot to float at this deepness.
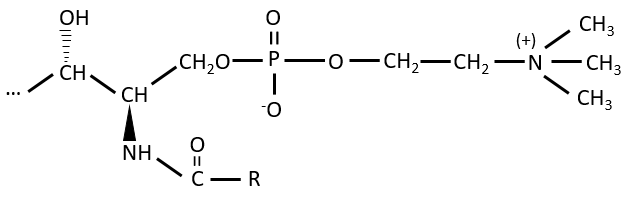
Polar lipids often present a phosphate or a charged amine
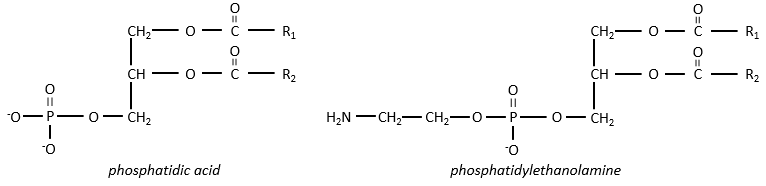
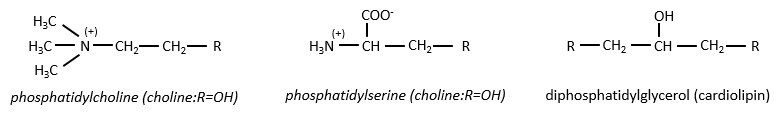
There are a bunch of derivates of the phosphatidic acid. Noting the phosphatidic acid R,
Many of them are constituting the membranes of cell. Oses can also be bound to lipids by via ester liaison and serve, in this case, as an element of signalisation.
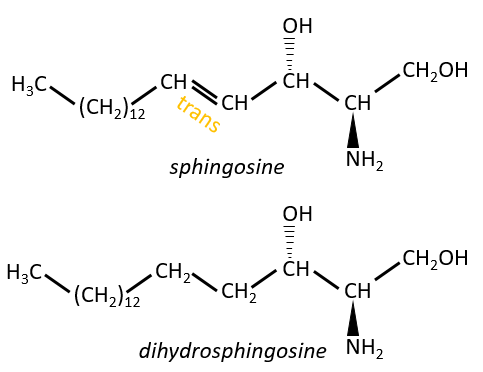
Sphingolipids are derivatives of the sphingosine and of the dihydrosphingosine.
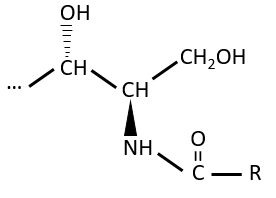
The lipids are bounds via an amide liaison.
Elements giving different properties to the molecule can bind on the alcohol. For instance, a choline can bind there with the help of a phosphodiester liaison.
This molecule is a sphingomyelin and is found in the membrane that surrounds some nerve cell axons. On the ester, an ose can be bound. Those molecules are called neutral glycosphingolipids or cerebrosides because they are often found in the membranes of the brain. Again, the sugar is an element of recognition/signalisation.
The ose can also be acid (for instance a sialic acid) and then the molecule is an acid glycosphingolipid, usually serving as receptor.
Non-saponifiable lipids
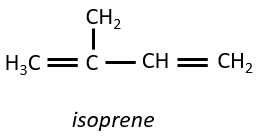
Terpenes
They are small molecules deriving from the isoprene.
The smallest terpenes are the monoterpenes, resulting from the combination of two isoprenes.
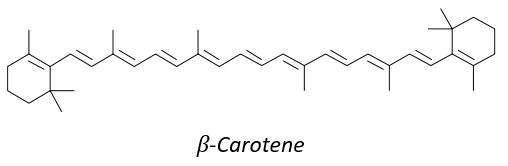
8 equivalents of the isoprene forms the beta-carothene. This molecule of 40 carbons is an antioxidant that protects against free radicals: it is easily excited because the electrons can be delocalised.
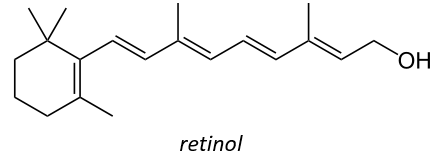
It is also a precursor of the vitamin A. It is a group of unsaturated nutritional organic compounds (found in carrots, amongst others) that includes retinol, retinal, retinoic acid, and several provitamin A carotenoids (most notably beta-carotene).
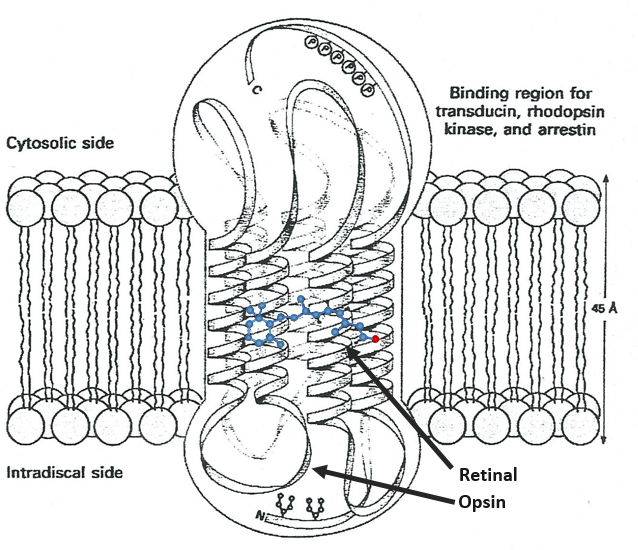
Vitamin A has multiple functions: it is important for growth and development, for the maintenance of the immune system and good vision. Vitamin A is needed by the retina of the eye in the form of retinal, which combines with protein opsin to form rhodopsin, the light-absorbing molecule necessary for both low-light (scotopic vision) and colour vision.
There are two types of captors in the eye: cones and rods. The first detect the colours and the seconds the light. The rods are composed of a series of bilipidic discs with rhodopsin receptors. Between the helixes of opsin is the 11-cis-retinal
All the liaisons except one in the retinal are trans. When a photon hits the retinal, this liaison becomes trans, perturbing the molecule and then the proteinic helixes that transfer the nervous signal. One photon is enough to generate a signal.
In the cones, there are specific modifications in the structure of the protein and the photon has to be in a specific range of wavelength (i.e. one colour) to transfer the signal.
Steroids
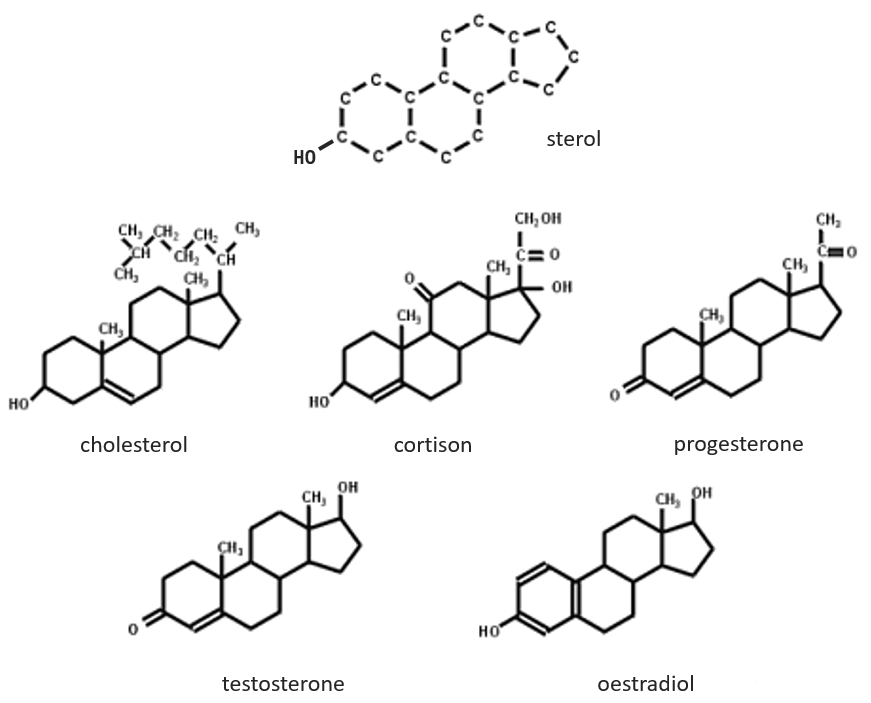
The base of steroids is the sterol, a molecule made of three cycles of 6 C and one of 5 C.
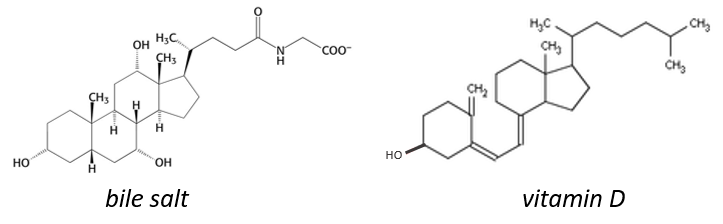
The cholesterol is completely hydrophobic. It is a precursor of several hormones, of the vitamin D and of the bile salt.
We often hear talking about the good and the bad cholesterol. It is actually an incorrect locution. The mode of transportation of the cholesterol is actually the important principle. The cholesterol is transported by lipoproteins (a combination of lipids and proteins) called LDL and HDL (for low and high density lipoproteins). The LDL transports cholesterol in the body from the liver. It is dropped off at some specific receptors to the LDL. If there is too much cholesterol, it will be dropped off anyway. The role of the HDL is to bring back the unused cholesterol back to the liver where it is destroyed. There is thus an equilibrium between the cholesterol that comes from and back to the liver.
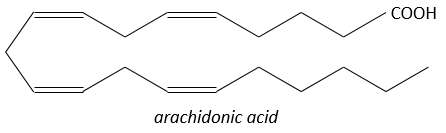
Prostaglandins
They are hormones acting on a short distance, with a short life time and they are all made from the arachidonate, one lipid of the cellular membrane, by a phospholipase. Prostaglandins activate numerous membrane receptors.
Following a given stimulus, some agents are activated to activate the PLA2 (phospholipase A2). The PLA2 cleaves phospholipids of the membrane to obtain fatty acids. From one fatty acid 4 types of prostaglandins are formed:
- prostacyclin: they are found in the wall of blood vessels and prevent the coagulation.
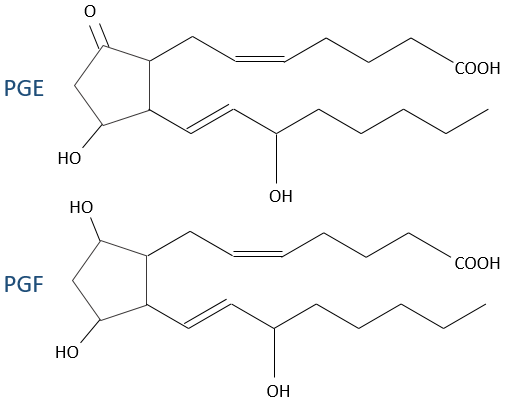
- PGE and PGF (stand for prostaglandin E and F): they are responsible of the sleep cycle and from the contractions at childbirth.
- thromboxane: they are implicated in the coagulation process.
- leukotrienes: we find them in the wall of the lungs. Asthmatics have an overproduction of leukotrienes.
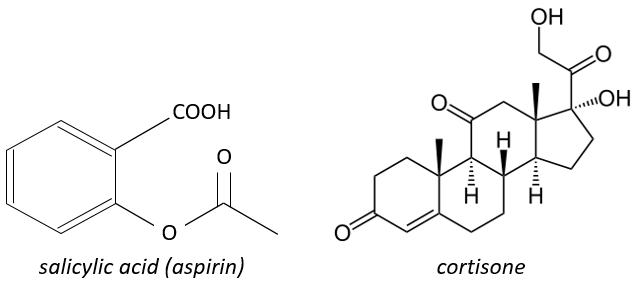
The aspirin blocks the activity/production of the cyclooxygenase which is an enzyme responsible in the formation of the four types of prostaglandins from the arachidonate. The cortisone blocks the PLA2.
Main function of the lipids: constituent of membranes
Membranes of cells separate the inside of the cell from the outside. Both sides are aqueous but the inside of the membrane is hydrophobic. Bilipidic membranes form spontaneously (called liposomes). Membranes are composed of lipids and of proteins with a composition that depends on the cell: basically the proteins allow exchanges and transports from one side of the membrane to the other side while the lipids are the concrete wall of the membrane. Cholesterol places itself in the hydrophobic part of the membrane to consolidate it.