The central dogma of molecular biology
The central dogma is the classic sequence of events :
DNA produces RNA by transcription and RNA produces proteins ( structural proteins and Enzymes ) by translation .
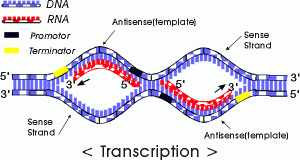
Transcription: making of RNA (taking the information of DNA and copying the information to (transitional form) RNA which is unstable.
Translation: is the act of synthesizing a protein from the information in the RNA.
Against central dogma:
-Enzymes can be also certain forms of RNA.
-Now we can take RNA and make DNA from that.
-There are enzymes that can make RNA from RNA.
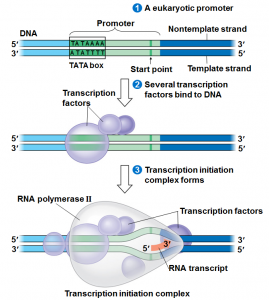
We will begin by transcription in prokaryotes.Typically in prokaryotes the transcription produces an RNA that encodes for more than one protein. Transcription starts at a promoter and make RNA at the same direction that DNA polymerase do for DNA (5’→3’) direction. RNA polymerase which recognizes promoter and does transcription (promoter is defined the site that bound by RNA polymerase and where transcription starts).
There are 2 parts to a promoter : there are so called -10 sequences and so called -35 sequences. What the -10 and -35 refer to is the distance in base pairs from the first base of DNA that is copied into RNA.These sequences are highly characteristic of all genes in the case of -10 of the classes and the genes on the -35 sequences.
The -35 sequence is recognized by a protein (a family of proteins referred to) as sigma factors, these are regulatory proteins. Bacterial RNA polymerases are formed by 4 different subunits (2 copies of big subunits called α and copies of β and β’) ⇒ α2ββ’.
α2ββ’ is the holoenzyme and then σ factor is a regulatory subunit and comes in many varieties. There are many σ factors ,the key thing with σ is that they are specific to particular promoters ,examples :
1)Heatshok
2) sporulation
There are many factors, the key thing with factors is that they are specific to particular promoters.
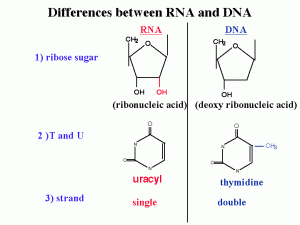
Differences between DNA and RNA :
1)2’ OH is special for RNA
DNA is deoxy. This OH group makes RNA unstable and vulnerable by enzymes or bases or different kinds of biophysical conditions. So RNA is designed to be unstable. Why RNA must be unstable (the earliest forms of life used RNA as a genetic material). Why we use an unstable molecule? probably it’s for the control of gene expression.
2) uracyl instead of thymidine
3) no primer required for the synthesis of RNA from RNA
4) synthesized in 5’→3’ direction (the chemistry involved is identical to DNA polymerase) :Initiation-elongation-termination
Step 1 :closed promoter complex
Step 2: formation of open promoter complex which is 17 bases pairs (normally there are 10 bases pairs by turn of helix) So 17 is almost 2 turns of helix in this situation, RNA polymerase start to synthesize RNA.
How we can confirm that? by DNA foot printing….